Being FAIR: Enabling Reproducible Data Science
- 1. Being FAIR: Enabling Reproducible Data Science Professor Carole Goble The University of Manchester, UK [email protected] 2018 Early Detection of Cancer Conference, OHSU, Portland, Oregon USA, 2-4 Oct 2018
- 2. Disclosure Knowledge management Computational workflows Sharing and exchange Reproducibility Large e-Infrastructure projects for life science data
- 3. The Learning Health System Phenotypic Patient Records Patient cohort building Patient stratification Case notes Discharge notes Patient cohorts Patient Multi-omics Public Reference repositories text mining, data mining data & vocabulary linking data analytics Single cell omics Clinical genomics Quantitative biology e-Health Predictive models Sensors Diagnostics Biomarkers Imaging Research Clinical Biobanks Scientific Literature Patient Public Health [Friedman]
- 4. An Inspiration http://fora.tv/2010/04/23/Sage_Commons_Josh_Sommer_Chordoma_Foundation Josh Sommer http://www.chordomafoundation.org/ Accelerate a cure Accelerate knowledge exchange
- 5. Barriers to Cure • Access to scientific resources • Coordination,Collaboration • Flow of Information • FAIR Data, FAIR Methods • FAIR Object Commons [Josh Sommer] GobleC., De Roure D., Bechhofer S. (2013) AcceleratingScientists’ KnowledgeTurns, https://doi.org/10.1007/978-3-642-37186-8_1 Research Commons Accelerate inter-lab knowledge turns Accumulate knowledge
- 6. 1. A Research Commons “… a “cloud-based” platform where investigators can store, share, access, and interact with digital objects (data, software, [models, SOPs], etc.) generated from …. research. By connecting the digital objects and making them accessible, the Data Commons is intended to allow novel scientific research that was not possible before, including hypothesis generation, discovery, and validation.” https://commonfund.nih.gov/commons Pooled Resources Federated Find andAccess Many entry points Data + Methods + Models
- 7. Clear steps Transparent Comprehensible Replicable Logged Accessible Provenance Standardised Harmonised Combined Method Materials Variations X N Repeat. Compare. Log & Track Provenance Scale 2. Data-driven Science, Predictive Science is Software-driven, Method-Driven
- 8. 3. Reuse and Reproducibility Is hard for in vivo/vitro and even for in silico analysis • OS version • Revision of scripts • Data analysis software versions • Version of data files • Command line parameters written on a napkin • “Magic” the grad student knows…. [Keiichiro Ono, Scripps Institute]
- 9. Findable (Citable) Accessible (Trackable) Interoperable (Intelligible) Reusable (Reproducible) Record Automate Contain Access
- 10. FAIR provenance portability preservation robustness access description standards, common APIs licensing standards, common metadata versioning, deviation variation sensitivity discrepancy handling parametric spaces packaging, containers dependencies steps ids Reproduce and reuse computations Transparently communicate the way computations are performed Disambiguate interpretation of inputs/parameters/results Safely (re)run computations ported onto different platforms Human and computer readable definitions for the provenance of computation, types for the data and results
- 11. Cancer Data Integrator [Várna,Davies, NIHR Health Informatics Collaborative, UK]
- 13. Objects: data + methods + models + provenance + Scharm M,Wendland F, Peters M,Wolfien M,TheileT,Waltemath D SEMS, University of Rostock zip-like file with a manifest & metadata - Bundle files - Keep provenance - Exchange data - Ship results Bergmann, F.T. (2014). COMBINE archive and OMEX format: one file to share all information to reproduce a modeling project. BMC bioinformatics,15(1), 1. Combine Archive Systems Biology Systems Medicine https://sems.unirostock.de/projects/combinearchive/
- 14. Research Object Framework Bechhofer et al (2013)Why linked data is not enough for scientists https://doi.org/10.1016/j.future.2011.08.004 Bechhofer et al (2010) Research Objects:Towards Exchange and Reuse of Digital Knowledge, https://eprints.soton.ac.uk/268555/ carry machine processable metadata in common and specific to different object types. bundle together and relate digital resources with their context into a unit. snapshot, cite, exchange run, evolve accumulate interlink Standards-based generic metadata framework
- 15. Container Metadata Object metadata, ontologies, identifiers “Unbounded” Objects Bags of things and external references to things Data used and results produced … Methods employed to produce and analyse that data … Provenance and settings … People involved … Annotations understanding & interpretation …
- 16. • Co-localizing massive genomics datasets, like The Cancer Genomics Atlas, alongside secure and scalable computational resources to analyze them. • Analyze own data alongside TCGA using predefined analytical workflows or your own tools. • Petabyte of multi- dimensional data available to authorized researchers. • Fully reproducible execution • Secure team collaboration. http://www.cancergenomicscloud.org/ NCI Cancer Genomics Cloud (CGC) Pilot
- 17. HTS pipelines for precision medicine GATK:Tumor-Normal Paired Exome-Sequencing pipeline [Durga Addepalli, Seven Bridges]
- 18. HTS pipelines for precision medicine GATK:Tumor-Normal Paired Exome-Sequencing pipeline [Durga Addepalli, Seven Bridges] Inputs OutputsAnalysis
- 19. Workflow Input Data (Files) Output Data (Files) Software Component Settings (Annotation) Workflow is defined usingCommonWorkflow Language (CWL) Software components are Docker images http://www.cancergenomicscloud.org/ Analysis
- 20. Output FilesInput Files Intermediates Parameters Configurations Workflow Run Provenance Narrative ExecutionWorkflow Engine Tools / Codes Resources Author Workflow Container Metadata Analysis
- 21. Parameters Configurations Workflow Provenance Workflow Engine Algorithms, Pipelines Definitions of the Metadata Instances Data files Computation metadata Tools / Codes metadata Biocompute workflow Data formats Ontologies Data files Results Container Stratified, Shareable Objects Scientifically reliable interpretation Verifiable results within acceptable uncertainty/error Comparable results
- 22. Parameters Configurations Workflow Provenance Workflow Engine Algorithms, Pipelines Definitions of the Metadata Instances Data files Computation metadata Tools / Codes metadata Biocompute workflow Data formats Ontologies Data files Results Container Biocontainers bio.tools CWLViewer
- 23. Open standards, commodity systems Describe and run workflows, and the command line tools they orchestrate, supporting containers to be portable, transparent and interoperable . Describe the workflow inputs, outputs, tools and data with controlled vocabularies / ontologies EDAM Describe the provenance of the workflow Software components are containerised to be portable Workflow systems run the CWL workflow Gathers the CWL workflow descriptions together with rich context and provenance using multi-tiered descriptions Snapshots the workflow. Relates it to other objects. Uses archive formats to contain the object
- 26. FAIR Methods, different workflow systems & clouds Living Products
- 27. https://osf.io/h59uh/ Personalized medicine regulation Standardize exchange of HTS workflows for regulatory submissions between FDA, pharma, bioinformatics platform providers and researchers Inspect and replicate the computational analytical workflow to review and approve the bioinformatics Domain-specific object model captures essential information without going in details of the actual execution. A community-driven project Emphasis on robust, safe reuse Technical Reproducibility packaging software and providing required datasets Human understanding of what has been done higher level steps of the workflow, their parameter spaces and algorithm settings Alterovitz, Dean II, Goble, Crusoe, Soiland-Reyes et al Enabling Precision Medicine via standard communication of NGS provenance, analysis, and results, biorxiv.org, 2017, https://doi.org/10.1101/191783
- 28. analysis and review sample archival sequencing run file transfer regulation computation pipelines produced files are massive in size transfer is slow too large to keep forever; not standardized difficult to validate/verify how can industry and FDA work together to avoid mistakes? HTS lifecycle: from a biological sample to biomedical research and regulation [Vahan Simonyan] FDA BAA contract HHSF223201510129C (PI: Raja Mazumder)
- 31. BioCompute Framework to advance Regulatory Science to support NGS analysis Emphasis on robust, safe reuse. Describe and validate the metadata of packages, and their contents, both inside and outside Standardise data formats and elements and exchange of Electronic Health Records Describe and validate analysis workflows, to be portable and interoperable Standardise and support sharing and analysis of Genomic data Ontologies Controlled vocabularies for describing all of the above APIs Programmable interfaces for accessing all of the above Alterovitz, Dean II, Goble, Crusoe, Soiland-Reyes et al Enabling Precision Medicine via standard communication of NGS provenance, analysis, and results, biorxiv.org, 2017, https://doi.org/10.1101/191783
- 33. Living Objectisms: grow, evolve, mutate • RO life cycles – Fixed snapshot – Living objects – Rot, mutate, clone • Arose from workflow sharing and preservation • Research Objects are analogous to software artefacts and practices rather than data or articles Snapshot Fork Combine
- 34. Validate Container Manifest Profile Descriptions what else is needed Dependencies Versioning its evolution what should be there Checklists Provenance where it came from ids metadata that describes Research Object general purpose to drive scalable infrastructure
- 35. All Type Specific Implementation specific Container Manifest Profile Descriptions what else is needed Dependencies Versioning its evolution what should be there Checklists Provenance where it came from ids metadata that describes Research Object
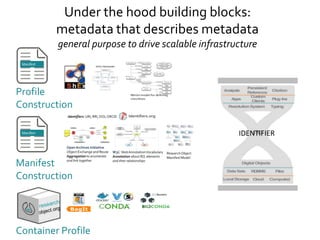
- 36. Container Profile Under the hood building blocks: metadata that describes metadata general purpose to drive scalable infrastructure Manifest Construction Profile Construction IDENTIFIER
- 37. Many other kinds of objects Multiple object types in an investigation Structured collections of objects Physical objects, SOPs These examples wereWorkflow Objects… [Sansone] Asthma Research e-Lab [Phil Crouch, John Ainsworth, Iain Buchan]
- 38. Chard et al: I'll take that to go: Big data bags and minimal identifiers for exchange of large, complex datasets, https://doi.org/10.1109/BigData.2016.7840618 Dnase HypersensitivityAnalysis using ENCODE (Encyclopedia of DNA Elements ) access, analysis and publishing using Galaxy images and genome sequences assembled from diverse repositories data distributed across multiple locations, referenced because big and persisted, efficiently and safely moved on demand Assemble and share large scale, multi-element datasets. [Chard, Kesselman, Foster, Madduri, 2016]
- 39. Richly structured descriptions of content in the bag and outside it Transfer and archive very large HTS datasets in a location- independent way. Secure referencing and moving of patient data. Big Data collections of arbitrary referenced content annotations, provenance, relations checksums Simple, location independent persistent identifiers Define a dataset and its contents by enumerating its elements, regardless of their location Verify and validate content
- 40. FAIR Data Commons 3. Everything is a research object: all the (distributed) components of an investigation (models, data, pipelines, SOPs, provenance...) into citable, exchangeable, publishable, preserved, nested objects 1. Assemble and share large scale, multi- element datasets. Secure referencing and moving of patient data. 2. Reproduce, port, share, and execute HTS pipelines (and other analytics …)
- 41. The Knowledge Object Reference Ontology (KORO): A formalism to support management and sharing of computable biomedical knowledge for learning health systems Flynn, Friedman, Boisvert, Landis‐Lewis, Lagoze (2018), https://doi.org/10.1002/lrh2.10054 Graphs of Research Objects Track Research Objects Combine and enrich Research Objects Learning Health Systems
- 42. International Efforts: FAIR Life Science Data Infrastructure • EGA in a Box for storing, coordinating and distributing human data • Human Data Beacons discovery service • Authentication and Authorization Infrastructure Interoperability, Compute, Data, Tools,Training Tools andWorkflow collaboratory for EOSC https://www.elixir-europe.org/use-cases/human-data
- 43. Summary: help knowledge turning • Data Science is underpinned by data access + transparent methods to enable reproducible and FAIR knowledge exchange. • FAIR First. • Research Objects as the currency of reproducibility and exchange • A bunch of tech, standards, tooling, best practices, grass roots and international activities going on. • Tech isn’t the issue. • e-Infrastructure matters. Please care about it.
- 45. Melissa Haendel, PhD Director of Translational Data Science, Oregon State University Director of the Center for Data to Health, Oregon Health & Science University
- 46. Acknowledgements Barend Mons Sean Bechhofer Matthew Gamble Raul Palma Jun Zhao Mark Robinson AlanWilliams Norman Morrison Stian Soiland-Reyes Tim Clark Alejandra Gonzalez-Beltran Philippe Rocca-Serra Ian Cottam Susanna Sansone KristianGarza Daniel Garijo Catarina Martins Iain Buchan Michael Crusoe Rob Finn Carl Kesselman Ian Foster Kyle Chard Vahan Simonyan Ravi Madduri Raja Mazumder GilAlterovitz, Denis Dean II Durga Addepalli Wouter Haak Anita De Waard Paul Groth Oscar Corcho Josh Sommer Project ID: 675728



![The Learning Health System
Phenotypic
Patient Records
Patient cohort building
Patient stratification
Case notes
Discharge notes
Patient cohorts
Patient Multi-omics
Public Reference
repositories
text mining, data mining
data & vocabulary linking
data analytics
Single cell omics
Clinical genomics
Quantitative biology
e-Health
Predictive
models
Sensors Diagnostics
Biomarkers
Imaging
Research Clinical
Biobanks
Scientific
Literature
Patient
Public Health
[Friedman]](https://image.slidesharecdn.com/goble-earlydetection-03102018-181015133246/85/Being-FAIR-Enabling-Reproducible-Data-Science-3-320.jpg)

![Barriers to Cure
• Access to scientific resources
• Coordination,Collaboration
• Flow of Information
• FAIR Data, FAIR Methods
• FAIR Object Commons
[Josh Sommer]
GobleC., De Roure D., Bechhofer S. (2013) AcceleratingScientists’ KnowledgeTurns, https://doi.org/10.1007/978-3-642-37186-8_1
Research Commons
Accelerate inter-lab
knowledge turns
Accumulate knowledge](https://image.slidesharecdn.com/goble-earlydetection-03102018-181015133246/85/Being-FAIR-Enabling-Reproducible-Data-Science-5-320.jpg)
![1. A Research Commons
“… a “cloud-based” platform where investigators can store, share, access, and interact
with digital objects (data, software, [models, SOPs], etc.) generated from …. research.
By connecting the digital objects and making them accessible, the Data Commons is
intended to allow novel scientific research that was not possible before, including
hypothesis generation, discovery, and validation.” https://commonfund.nih.gov/commons
Pooled Resources
Federated
Find andAccess
Many entry points
Data + Methods + Models](https://image.slidesharecdn.com/goble-earlydetection-03102018-181015133246/85/Being-FAIR-Enabling-Reproducible-Data-Science-6-320.jpg)

![3. Reuse and Reproducibility
Is hard for in vivo/vitro and
even for in silico analysis
• OS version
• Revision of scripts
• Data analysis software
versions
• Version of data files
• Command line parameters
written on a napkin
• “Magic” the grad student
knows….
[Keiichiro Ono, Scripps Institute]](https://image.slidesharecdn.com/goble-earlydetection-03102018-181015133246/85/Being-FAIR-Enabling-Reproducible-Data-Science-8-320.jpg)


![Cancer Data Integrator
[Várna,Davies, NIHR Health Informatics Collaborative, UK]](https://image.slidesharecdn.com/goble-earlydetection-03102018-181015133246/85/Being-FAIR-Enabling-Reproducible-Data-Science-11-320.jpg)





![HTS pipelines for precision medicine
GATK:Tumor-Normal Paired Exome-Sequencing pipeline
[Durga Addepalli, Seven Bridges]](https://image.slidesharecdn.com/goble-earlydetection-03102018-181015133246/85/Being-FAIR-Enabling-Reproducible-Data-Science-17-320.jpg)
![HTS pipelines for precision medicine
GATK:Tumor-Normal Paired Exome-Sequencing pipeline
[Durga Addepalli, Seven Bridges]
Inputs OutputsAnalysis](https://image.slidesharecdn.com/goble-earlydetection-03102018-181015133246/85/Being-FAIR-Enabling-Reproducible-Data-Science-18-320.jpg)









![analysis
and review
sample
archival
sequencing run
file transfer
regulation
computation
pipelines
produced files
are massive in
size
transfer is
slow
too large to keep
forever; not
standardized
difficult to
validate/verify
how can
industry and
FDA work
together to
avoid
mistakes?
HTS lifecycle: from a biological sample
to biomedical research and regulation
[Vahan Simonyan] FDA BAA contract HHSF223201510129C (PI: Raja Mazumder)](https://image.slidesharecdn.com/goble-earlydetection-03102018-181015133246/85/Being-FAIR-Enabling-Reproducible-Data-Science-28-320.jpg)







![Many other kinds of objects
Multiple object types in an
investigation
Structured collections of objects
Physical objects, SOPs
These examples wereWorkflow Objects…
[Sansone]
Asthma Research e-Lab
[Phil Crouch, John
Ainsworth, Iain Buchan]](https://image.slidesharecdn.com/goble-earlydetection-03102018-181015133246/85/Being-FAIR-Enabling-Reproducible-Data-Science-37-320.jpg)
![Chard et al: I'll take that to go: Big data bags and minimal identifiers for exchange of large, complex datasets, https://doi.org/10.1109/BigData.2016.7840618
Dnase HypersensitivityAnalysis
using ENCODE (Encyclopedia of
DNA Elements ) access, analysis
and publishing using Galaxy
images and
genome sequences
assembled from diverse
repositories
data distributed across
multiple locations,
referenced because big
and persisted, efficiently
and safely moved on
demand
Assemble and share large scale, multi-element
datasets.
[Chard, Kesselman, Foster, Madduri, 2016]](https://image.slidesharecdn.com/goble-earlydetection-03102018-181015133246/85/Being-FAIR-Enabling-Reproducible-Data-Science-38-320.jpg)







