BioThings API: Building a FAIR API Ecosystem for Biomedical Knowledge
- 1. Chunlei Wu, Ph.D. [email protected] @chunleiwu https://wulab.io Associate Professor Dept. of Integrative Structural and Computational Biology The Scripps Research Institute La Jolla, CA, USA 01/16/2019 NCI – CBIIT Speaker Series Building a FAIR API Ecosystem for Biomedical Knowledge http://biothings.io
- 2. Biomedical Data API API – Application Programming Interface API is a way to abstract the data-access layer.
- 3. APIs as a reusable data layer Presentation Layer Business logic Layer Data Layer Application 1 Presentation Layer Business logic Layer Data Layer Application 2 View Controller Model Repetitive data wrangling: • Parsing dump files • ID conversion • Data merging • Data transformation • Source monitoring • Download scheduler • … … Presentation Layer Business logic Layer Common Data Layer Application 1 Presentation Layer Business logic Layer Data Layer Application 2
- 4. Why bioinformaticians need APIs It's about Modularization photo credits: http://www.edmentum.com/sites/edmentum.com/files/solutions/content/building_0.jpg http://www.howcsharp.com/img/0/68/dont-repeat-yourself-dry-300x211.jpg http://blog.capinc.com/wp-content/uploads/2013/02/Recycle_Logo_by_Har1-300x263.png Reusability DRY principle
- 5. Biomedical APIs and FAIR matrix APIs are not quite findable APIs are naturally accessible But enterprise-grade Biomedical APIs are still few Often not interoperable across APIs APIs serve reusable piece of data But more can be made reusable in API development ? ?
- 6. Computer science is all about “Abstraction” “Abstraction” is the simple guiding-principle for informaticians Reducing repetitive efforts Opportunities for informaticians
- 7. An example: abstracting the gene search box http://biogps.org
- 9. Aggregated Gene annotations represented in JSON documents { “_id”: “1017”, “symbol”: “CDK2”, “ensembl”: “ENSG00000123374”, “refseq”: [ “NM_001798”, “NM_052827” ], “reporter”: { “U95A”: [ “1792_g_at”, “1833_at” ], “U133A”:[ “211804_s_at”, “2045252_at”, “211803_at” ] } } Source merging criteria: matching NCBI or Ensembl Gene ids HGNC MGI RGD Refseq Ensembl UniProt UniGene Homologene PantherDB GO Reactome Wikipathways KEGG PDB PFAM Interpro Prosite PIR Pharmgkb UMLS Wikipedia Pharos …
- 10. Gene-centric API via a simple interface Get gene object(s) via either NCBI/Ensembl gene ids: http://mygene.info/v3/gene/1017 http://mygene.info/v3/gene/ENSG00000123374 http://mygene.info/v3/gene/1017?fields=symbol,name,pathway,uniprot Find matching gene objects with any query terms: http://mygene.info/v3/query?q=CDK2 http://mygene.info/v3/query?q=name:kinase&species=human http://mygene.info/v3/query?q=name:kinase AND _exists_:pathway http://mygene.info/v3/query?q=pathway.kegg.name:wnt&fields=entrezgene,symbol,taxid,interpro Batch queries supported via POST
- 11. MyVariant.info API { "_id": "chr1:g.196659237C>T", "cosmic": { "chrom": "1", "hg19": { "start": 196659237, "end": 196659237 }, "ref": "C", "alt": "T", "tumor_site": "breast", "mut_freq": 0.49, "mut_nt": "C>T", "cosmic_id": "COSM424915" } { "_id": "chr1:g.196659237C>T", "cadd": { … }, "clinvar": { … }, "cosmic": { … }, "dbsnp": { … }, "dbnsfp": { … }, "evs": { … }, "emv": { … }, "mutdb": { … }, "gwassnp": { … }, "snpedia": { … }, "wellderly": { … } } Source merging criteria: matching HGVS names Only genomic-based HGVS names are used (support both hg19 and hg38) more at: http://docs.myvariant.info/en/latest/doc/data.html#id-field http://myvariant.info A real example online 21 sources: dbSNP dbNSFP CADD UniProt ClinVar CIVIC CGI DOCM ExAC GNOMAD EMV EVS Grasp SNPEFF …
- 12. MyVariant.info API Data source license and metadata: { "_id": "chr1:g.196659237C>T", "cadd": { "_license": “http://bit.ly/2TIuab9”, … }, "clinvar": { "_license": “http://bit.ly/2SQdcI0”, … }, " civic": { "_license": “http://bit.ly/2FqS871”, … }, “dbnsfp": { "_license": “http://bit.ly/2VLnQBz” , … }, … } { "build_date": "2018-12-06T22:15:39.743302", "build_version": "20181206", "src": { "cadd": { "license_url": "http://cadd.gs.washington.edu/contact", "license_url_short": "http://bit.ly/2TIuab9", "stats": { "cadd": 226932858 }, "url": "http://cadd.gs.washington.edu/home", "version": "1.3" }, "civic": { "licence": "CC0 1.0 Universal", "license_url": "https://creativecommons.org/publicdomain/zero/1.0/", "license_url_short": "http://bit.ly/2FqS871", "stats": { "civic": 1559 }, "url": "https://civicdb.org", "version": "201706" }, … }} “_license” urls embedded in every response Detailed source metadata at http://myvariant.info/metadata
- 13. MyChem.info API for chemicals and drugs { "_id": "RRUDCFGSUDOHDG-UHFFFAOYSA-N", “chebi": { “id”: “CHEBI:49029”, “formulae”: “C2H5NO2", “name”: “N-hydroxyacetimidic acid”, “smiles”: “CC(O)=NO”, “xrefs": { “pubchem": { “cid”: “1990”, “sid”: “49693671” } } }, “drugbank”: {…}, “drugcentral”: {…} } Source merging criteria: matching InChiKey more at: http://docs.mychem.info/en/latest/doc/data.html#id-field 11 sources: AEOLUS ChEBI ChEMBL Drugbank Drugcentral GINAS NDC PharmGKB PubChem UNII
- 14. Collectively, we call them “BioThings APIs” Aggregates annotations for 96 million drugs/chemicals from 11 resources I have a list of drug/chemical ids, want to get annotations about them? Drug/chemical annotation service: GET /v1/drug/<drugid> POST /v1/drug/ (batch mode) I want to get matching drugs/chemicals with my query term(s) Drug/chemical query service: GET /v1/query/?q= <query> POST /v1/query/ (batch mode) http://mygene.info http://myvariant.info http://mychem.info ~10 M requests ~20,000 unique IPs every month ~5 M requests 8000 unique IPs every month recently launched! Aggregates annotations for 25 million genes from 30 resources I have a list of gene ids, want to get annotations about them? Gene annotation service: GET /v3/gene/<geneid> POST /v3/gene/ (batch mode) I want to get matching genes with my query term(s) Gene query service: GET /v3/query/?q= <query> POST /v3/query/ (batch mode) Aggregates annotations for 874 million variants from 21 resources I have a list of variant ids, want to get annotations about them? Variant annotation service: GET /v1/variant/<hgvsid> POST /v1/variant/ (batch mode) I want to get matching variants with my query term(s) Variant query service: GET /v1/query/?q= <query> POST /v1/query/ (batch mode)
- 15. Who is using BioThings API Many users use our APIs in their daily analysis pipelines or simply caching annotations locally http://biothings.io/who-is-using
- 16. Who is using BioThings API Baylor College of Med 17,264,902 OHSU 16,442,387 Google LLC 590,305 UNC 480,168 Cincinnati Children 229,686 Université Laval 226,243 UCSD 101,867 Rockefeller University 96,018 Illumina 92,902 Yale Univ 44,587 NY Genome Center 3,502,635 UTexas-Austin 2,785,542 Stanford University 2,607,072 Univ of Colorado 1,325,650 Yale Univ 1,054,124 Vanderbilt Univ 851,375 Univ of Chicago 614,891 Baylor College of Med 550,022 Oregon State Univ 525,350 Univ of Illinois - UC 507,421 Top 10 organizations* and their requests (01/01/2018-12/31/2018) * Orgs mapped to the general ISPs were removed # of requests # of requests
- 17. BioThings API usage by numbers Total requests 130M Avg. Monthly requests 10.7M Total Unique IPs 173K Monthly Unique IPs ~19K mygene Python client monthly download ~4470 mygene R client monthly download ~611 Availability tracked by UptimeRobot 100% Based on usage data (01/01/2018-12/31/2018) Total requests 55M Average Monthly requests 4.6M Total Unique IPs 86K Monthly Unique IPs ~8K myvariant Python client monthly download ~3600 myvariant R client monthly download ~164 Availability tracked by UptimeRobot 100%
- 18. mygene and myvariant Python clients Open source repositories depending on our python clients (total 29) (total 11) https://libraries.io/pypi/mygene https://libraries.io/pypi/myvariant
- 19. Build Enterprise-grade Biomedical APIs Simple to use Always up-to-date (weekly updated) Comprehensive - MyGene.info: 25M genes from 24K species - MyVariant.info: 874M (700M observed) - MyChem.info: 96M chemicals/drugs High-performance and scalable High-availability Python, R, JavaScript clients Developer-friendly (support CORS, gzip, https, msgpack, etc.) • “fetch_all” feature for streaming large query results
- 20. A collection of high- performance APIs http://T.biothings.io fast, up-to-date, simple-to-use Gene Variant Drug/Chemical Taxonomy http://MyDisease.info Disease What about other “BioThings”, with our limited bandwidth? Can we further abstract the process of making APIs? Help ourselves as well as others to build APIs.
- 21. Schematic view of MyVariant.info architecture Web module Hub module Individual server node * Colors indicate the different updating schedules
- 22. Others can build their own APIs with src monitor scheduler data merger data indexer URL pattern JSONP CORS compression JSON-LD Tracking unit tests cluster setup data deploy cluster scaling load-balancing Optional query customization Data Hub Web API Cloud Deployment data parsers for individual resources MongoDB + Elasticsearch Python/Tornado Amazon AWS http://docs.biothings.io BioThingsSDK done by Users abstracted in SDK
- 23. My data file I will write a parser Describe data schema for indexing Setup Elasticsearch Index JSON objects in Elasticsearch Ready to serve Your BioThings API is live! LIVE Inspector indexer In [1]: from biothings.www import BiothingsAPIApp In [2]: drug_api_app = BiothingsAPIApp( ...: APP_LIST= [(r'/v1/drug/(.+)/?', 'BiothingHandler'), ...: (r'/v1/drug/?$', 'BiothingHandler')], ...: ES_INDEX=‘drug_databuild_20170708', ES_DOC_TYPE=‘drug') In [3]: drug_api_app.start(port=8002) INFO:root:Server is running on "0.0.0.0:8002"... code snippet user actions done by SDK Scenario 1 - I have a data file, and I want to make it an API: - Turn a data file into a high-quality API http://docs.biothings.io/en/latest/doc/single_source_tutorial.html
- 24. - Unified API clients in Python/R/JS # Access your live API from the unified Python client: In [1]: from biothings_client import get_client In [2]: mydrug = get_client("drug", url="localhost:8002/v1") In [3]: mydrug.getdrug("DB08571”) In [4]: mydrug.query("drugbank.name:celecoxib") In [5]: mygene = get_client("gene") In [6]: mygene.getgene("1017") In [7]: mygene.query("symbol:cdk2") In [8]: myvariant = get_client("variant“) In [9]: myvariant.getvariant("chr7:g.140453134T>C") In [10]:myvariant.query("dbsnp.rsid:rs58991260") User API MyGene.info API MyVariant.info API biothings_client available in Python R Javascript https://biothings-clientpy.readthedocs.io
- 25. - Merging and keeping data sources in-sync Scenario 2 - I need to aggregate multiple data sources, and keep them up-to-date: A data source management console included in SDK http://docs.biothings.io/en/latest/doc/hub_tutorial.html
- 26. BioThings Studio as web-based development environment Contribute to the existing BioThings APIs Build your own API Biomedical Data Sources (MyGene.info data sources shown in BioThings Studio) https://github.com/biothings/biothings_studio
- 27. What about data schemas? BioThings API and SDK are data-schema neutral, but can be customized to be an specialized API and SDK focusing on a particular schema or vocabulary standards. Schemas Ontologies Vocabularies Specialized API and SDK Incentivize the adoption of standards
- 28. A collection of high- performance APIs An SDK for building your own APIs http://T.biothings.io fast, up-to-date, simple-to-use JSON data aggregation mechanism High- performance query engine Well-designed REST API pattern JSON-LD enabled Linked Data Data-updating scheduler Python/R clients … Your data source Your API Abstraction of API building/deployment Gene Variant Drug/Chemical Taxonomy http://MyDisease.info Disease What about other APIs? How can APIs work together?
- 29. Use cases in NCATS Translator Program NCATS Biomedical Data Translator Program https://ncats.nih.gov/translator Two proof-of-concept queries For each of the drug-condition pairs listed below, construct a clinical outcome pathway that best explains how the drug effects its action. Drug Condition METADOXINE Hepatitis, Alcoholic MEMANTINE Alzheimer Disease OXYMORPHONE Anxiety … … For each of the diseases listed below, list which other genetic conditions observed in the human population might offer protection AND WHY. Disease Osteoporosis Asthma Ebola Virus Infection …
- 30. API-level data integration for translational research Electronic Health Record (EHS) Drugs Proteins Pathways Genes Variants MyVariant.info ClinVar CiVIC … MyGene.info Ensembl … Reactome WikiPathways …UniProt … MyChem.info Clue.io DrugBank … Pharos Biolink Wikidata NDEx …
- 32. Input Output 1. Compacted Format 2. Compacted Format 3. Nquads Format Semantically-aligned API output The separation of data and its semantic context: • Deal with data first, and semantic second • Deal with data only and others can help the semantic annotations
- 33. Semantic relationship represented in JSON-LD { "_id": "RZVAJINKPMORJF-UHFFFAOYSA-N", "indication":[ { concept_id: "Migraine", concept_name: "37796009" }, ... ] } { "@context": { "indication": { "@type": "@id", "@id": "assoc:treats", "@context": { "concept_name": { "@type": "@id", "@id": "attr:label", "@context": { "@base": "http://biothings.io/explorer/vocab/terms/disease-name/" } }, "concept_id": { "@type": "@id", "@id": "attr:id", "@context": { "@base": "http://identifiers.org/snomedct/" } } } } } } acetaminophen Migraine treats JSON object JSON-LD context
- 34. OpenAPI specifications for API metadata Tells how an API works
- 35. SmartAPI built on community standards http://smart-api.info Adds the semantic context for the data served from an API Tells how an API works
- 36. SmartAPI defines extensions for rich API metadata Biological domain-specific metadata fields
- 37. SmartAPI as an API registry http://smart-api.info
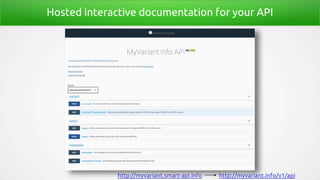
- 38. Hosted interactive documentation for your API http://myvariant.smart-api.info http://myvariant.info/v1/api
- 39. Project-specific API portals https://smart-api.info/registry/translatorhttps://smart-api.info/registry/nihdatacommons NIH Data Commons Project NCATS Translator Project
- 40. A Real-world Translational Questions From NCATS Translator Hackathon in May 2018 Disease - Gene Gene - Pathways Pathways - GeneGene - Chemical Symptom - Disease
- 41. To explore the network of “SmartAPIs”: http://biothings.io/explorer/ http://biothings.io/explorer_beta/ Discover APIs for specific tasks Automatically trigger API calls to construct a subset of the knowledge graph Downstream analysis
- 42. Find APIs can get me from pathways to genes: Pathways Available APIs Genes biocarta kegg wikipathway reactome ncbigene uniprot
- 43. Find associated drug compounds to gene LCK: LCK CHEML3707348 LCK inhibits Via DGIDB API INCHIKEY:KKYYLKPGILUPOA-UHFFFAOYSA-N UniProt:P06239 equals Via MyGene API targets Via MyChem API CHEMBL223873 equals Via MyChem API
- 44. More about Video Tutorial https://youtu.be/cPUKRsaTlhg BioThings Explorer API: http://biothings.io/explorer/api/ Demos in Jupyter Notebook: BioThings Explorer Demo BioThings Explorer Metadata http://biothings.io/explorer/
- 45. BioThings project as a FAIR API Ecosystem Accessible Findable Interoperable Reusable If you want fast and update- to-date access to gene, variant, chemical, drug data. If you want to quickly turn your data into an high- performance API. If you built your API and want others to find your API and use it together with other APIs for a specific workflow.
- 46. Acknowledgement Scripps Research Andrew Su (sulab.org) Cyrus Afrasiabi Sebastien Lelong Jiwen (Kevin) Xin Marco Cano Alvarado Ginger Tsueng Byung Ryul Jeon Greg Taylor Xinhua (Jerry) Zhou Nina Moore Maastricht Univ. Michel Dumontier (dumontierlab.com) Amrapali Zaveri Kody Moodley Trish Whetzel (EBI) Shima Dastgheib (NuMedii) Ruben Verborgh (Ghent Univ.) Paul Avillach (Harvard) Gabor Korodi (Harvard) Raymond Terryn (Univ. of Miami) Kathleen Jagodnik (Mount Sinai) Pedro Assis (Stanford) Funding support from NIH Data Commons API interoperability working group Univ. of Washington Sean Mooney Vikas R Pejaver Translator, CD2H
Editor's Notes
- #11: Up-to-date and high-performance and high-availability








![Aggregated Gene annotations represented in JSON documents
{
“_id”: “1017”,
“symbol”: “CDK2”,
“ensembl”: “ENSG00000123374”,
“refseq”: [
“NM_001798”,
“NM_052827”
],
“reporter”: {
“U95A”: [
“1792_g_at”,
“1833_at”
],
“U133A”:[
“211804_s_at”,
“2045252_at”,
“211803_at”
]
}
}
Source merging criteria:
matching NCBI or Ensembl Gene ids
HGNC
MGI
RGD
Refseq
Ensembl
UniProt
UniGene
Homologene
PantherDB
GO
Reactome
Wikipathways
KEGG
PDB
PFAM
Interpro
Prosite
PIR
Pharmgkb
UMLS
Wikipedia
Pharos
…](https://image.slidesharecdn.com/biothingsncicbiit201901-190130191603/85/BioThings-API-Building-a-FAIR-API-Ecosystem-for-Biomedical-Knowledge-9-320.jpg)













![My data file
I will write a
parser
Describe data
schema for
indexing
Setup
Elasticsearch
Index JSON
objects in
Elasticsearch
Ready to
serve
Your BioThings
API is live!
LIVE
Inspector
indexer
In [1]: from biothings.www import BiothingsAPIApp
In [2]: drug_api_app = BiothingsAPIApp(
...: APP_LIST= [(r'/v1/drug/(.+)/?', 'BiothingHandler'),
...: (r'/v1/drug/?$', 'BiothingHandler')],
...: ES_INDEX=‘drug_databuild_20170708', ES_DOC_TYPE=‘drug')
In [3]: drug_api_app.start(port=8002)
INFO:root:Server is running on "0.0.0.0:8002"...
code snippet
user actions
done by SDK
Scenario 1 - I have a data file, and I want to make it an API:
- Turn a data file into a high-quality API
http://docs.biothings.io/en/latest/doc/single_source_tutorial.html](https://image.slidesharecdn.com/biothingsncicbiit201901-190130191603/85/BioThings-API-Building-a-FAIR-API-Ecosystem-for-Biomedical-Knowledge-23-320.jpg)
![- Unified API clients in Python/R/JS
# Access your live API from the unified Python client:
In [1]: from biothings_client import get_client
In [2]: mydrug = get_client("drug", url="localhost:8002/v1")
In [3]: mydrug.getdrug("DB08571”)
In [4]: mydrug.query("drugbank.name:celecoxib")
In [5]: mygene = get_client("gene")
In [6]: mygene.getgene("1017")
In [7]: mygene.query("symbol:cdk2")
In [8]: myvariant = get_client("variant“)
In [9]: myvariant.getvariant("chr7:g.140453134T>C")
In [10]:myvariant.query("dbsnp.rsid:rs58991260")
User API
MyGene.info API
MyVariant.info API
biothings_client available in
Python R Javascript https://biothings-clientpy.readthedocs.io](https://image.slidesharecdn.com/biothingsncicbiit201901-190130191603/85/BioThings-API-Building-a-FAIR-API-Ecosystem-for-Biomedical-Knowledge-24-320.jpg)








![Semantic relationship represented in JSON-LD
{
"_id": "RZVAJINKPMORJF-UHFFFAOYSA-N",
"indication":[
{
concept_id: "Migraine",
concept_name: "37796009"
},
...
]
}
{
"@context": {
"indication": {
"@type": "@id",
"@id": "assoc:treats",
"@context": {
"concept_name": {
"@type": "@id",
"@id": "attr:label",
"@context": {
"@base": "http://biothings.io/explorer/vocab/terms/disease-name/"
}
},
"concept_id": {
"@type": "@id",
"@id": "attr:id",
"@context": {
"@base": "http://identifiers.org/snomedct/"
}
}
}
}
}
}
acetaminophen Migraine
treats
JSON object
JSON-LD context](https://image.slidesharecdn.com/biothingsncicbiit201901-190130191603/85/BioThings-API-Building-a-FAIR-API-Ecosystem-for-Biomedical-Knowledge-33-320.jpg)