Ad
Data cleaning with the Kurator toolkit: Bridging the gap between conventional scripting and high-performance workflow automation
- 1. Data cleaning with the Kurator toolkit Bridging the gap between conventional scripting and high-performance workflow automation Timothy McPhillips, David Lowery, James Hanken, Bertram Ludäscher, James A. Macklin, Paul J. Morris, Robert A. Morris, Tianhong Song, and John Wieczorek TDWG 2015 - Biodiversity Informatics Services and Workflows Symposium Nairobi, Kenya - September 30, 2015
- 2. Kurator: workflow automation for cleaning biodiversity data Project aims § Facilitate cleaning of biodiversity data. § Support both traditional scripting and high-performance scientific workflows. § Deliver much more than a fixed set of configurable workflows. Technical strategy § Wrap Akka actor toolkit in a curation-oriented scientific workflow language and runtime. § Enable scientists who write scripts to add their own new data validation and cleaning actors. § Bring to scripts major advantages of workflow automation: prospective and retrospective provenance. § Bridge gaps between data validation services, data cleaning scripts, and pipelined data curation workflows. Empower users and developers of scripts, actors, and workflows.
- 3. What some of us think of when we hear the term ‘scientific workflows’ Phylogenetics workflow in Kepler (2005) Graphical interface § Canvas for assembling and displaying the workflow. § Library of workflow blocks (‘actors’) that can be dragged onto the canvas and connected. § Arrows that represent control dependencies or paths of data flow. § A run button. These features are not essential to managing actual scientific workflows.
- 4. 10 essential functions of a scientific workflow system 1. Automate programs and services scientists already use. 2. Schedule invocations of programs and services correctly and efficiently – in parallel where possible. 3. Manage data flow to, from, and between programs and services. 4. Enable scientists (not just developers) to author or modify workflows easily. 5. Predict what a workflow will do when executed: prospective provenance. 6. Record what actually happens during workflow execution. 7. Reveal retrospective provenance – how workflow products were derived from inputs via programs and services. 8. Organize intermediate and final data products as desired by users. 9. Enable scientists to version, share and publish their workflows. 10. Empower scientists who wish to automate additional programs and services themselves. These functions–not actors—distinguish scientific workflow automation from general scientific software development.
- 5. Why build yet another system? Available systems § Kepler (Ptolemy II), Taverna, VisTrails… § Familiar graphical programming environments. Limitations § Often little support for organizing intermediate and final data products in ways familiar to scientists. § Professional software developers frequently are needed to develop new components or workflows. Huge gap between how these systems are used and how scientists already automate their analyses—via scripting. Part of a Kepler workflow for inferring phylogenetic trees from protein sequences.
- 6. Avoiding actor ‘overuse injuries’ Overuse the actor paradigm… § In many systems workflows can be reused as actors, or ‘subworkflows’ in other workflows. § This is a necessary abstraction when workflow systems are not well-integrated with scripting languages. § Each actor at left is a page of Java code. But this whole ‘subworkflow’ could be written in a half a page of Python! …or use the right tools for the right job! § In Kurator we want to enable the actor abstraction where it pays off the most—as the unit of parallelism. § For specifying behavior inside of actors why not use easy to understand scripts? New actors and workflows must be easy and fast to develop. Part of a Kepler workflow for inferring phylogenetic trees from protein sequences.
- 7. Data curation workflow using Kepler FilteredPush explored using workflows for data cleaning § First used COMAD workflow model supported by Kepler. § Enabled graphical assembly and configuration of custom workflows from library of actors. Highlighted potential performance limitations of workflow engines.
- 8. FP-Akka workflows load data check scientific name check basis of record check date collected check lat/long write out results § FilteredPush next investigated use of the Akka actor toolkit and platform. § Widely used in industry, well supported, and rapidly advancing. § Efficient parallel execution across multiple processors and compute nodes. § Improved performance and scalability compared to Kepler. Limitations of directly using Akka § Writing new Akka actors and programs requires Java (or Scala) experience. § Must address many parallel programming challenges from scratch. Advanced programming skills required to write Akka programs (‘workflows’) that run correctly.
- 9. Akka partly supports two essential workflow platform requirements The Kurator toolkit will satisfy the rest as needed. 1. Automate programs and services scientists already use. 2. Schedule invocations of programs and services correctly and efficiently–in parallel where possible. 3. Manage data flow to, from, and between programs and services. 4. Enable scientists (not just developers) to author or modify workflows easily. 5. Predict what a workflow will do when executed: prospective provenance. 6. Record what actually happens during workflow execution. 7. Reveal retrospective provenance – how workflow products were derived from inputs via programs and services. 8. Organize intermediate and final data products as desired by users. 9. Enable scientists to version, share and publish their workflows. 10. Empower scientists who wish to automate additional programs and services themselves.
- 10. The Kurator Toolkit YesWorkflow (YW) § Add YW annotations to any script or program that supports text comments. Highlight the workflow structure in the script. § Visualize or query prospective provenance before running a script. § Reconstruct, visualize, and query retrospective provenance after running the script. § Integrate provenance gathered from file names and paths, log files, data file headers, run metadata, and records of run-time events. Kurator-Akka § Write functions or classes in Python or Java. Mark up with YesWorkflow. § Declare how scripts or Java code can be used as actors. Short YAML blocks. § Declare workflows. List actors and specify their connections. More YAML. § Run workflow. Use Akka for parallelization transparently—and correctly. § Reconstruct retrospective provenance of workflow products.
- 11. Example: data validation and cleaning using WoRMS web services 1) Write simple Python functions (or a class) that wrap the web services provided by the World Register of Marine Species (WoRMS). 2) Develop a Python script that uses the service wrapper functions from (1) to clean a set of records provided in CSV format. 3) Mark up the script written in (2) with YesWorkflow annotations and graphically display the script as a workflow. 4) Factor out of (2) a Python function that can clean a single record using the WoRMS wrapper functions in (1). 5) Write a block of YAML that declares how the record-cleaning function in (4) can be used as an actor in a Kurator-Akka workflow. 6) Declare using YAML a workflow that uses the actor declared in (5) along with actors for reading and writing CSV files. 7) Run the workflow (6) on a sample data set with the CSV reader, CSV writer and WoRMS validation actors all running in parallel. README at https://github.com/kurator-org/kurator-validation shows how to: Illustrates how Kurator aims to facilitate composition of actors and high-performance workflows from simple functions and scripts.
- 12. class WoRMSService(object): """ Class for accessing the WoRMS taxonomic name database via the AphiaNameService. The Aphia names services are described at http://marinespecies.org/aphia.php?p=soap. """ WORMS_APHIA_NAME_SERVICE_URL = 'http://marinespecies.org/aphia.php?p=soap&wsdl=1’ def __init__(self, marine_only=False): """ Initialize a SOAP client using the WSDL for the WoRMS Aphia names service""" self._client = Client(self.WORMS_APHIA_NAME_SERVICE_URL) self._marine_only = marine_only def aphia_record_by_exact_taxon_name(self, name): """ Perform an exact match on the input name against the taxon names in WoRMS. This function first invokes an Aphia names service to lookup the Aphia ID for the taxon name. If exactly one match is returned, this function retrieves the Aphia record for that ID and returns it. """ aphia_id = self._client.service.getAphiaID(name, self._marine_only); if aphia_id is None or aphia_id == -‐999: # -‐999 indicates multiple matches return None else: return self._client.service.getAphiaRecordByID(aphia_id) def aphia_record_by_fuzzy_taxon_name(self, name): : : Python class wrapping WoRMS services WoRMS services
- 13. # @BEGIN find_matching_worms_record # @IN original_scientific_name # @OUT matching_worms_record # @OUT worms_lsid worms_match_result = None worms_lsid = None # first try exact match of the scientific name against WoRMS matching_worms_record = worms.aphia_record_by_exact_taxon_name(original_scientific_name) if matching_worms_record is not None: worms_match_result = 'exact' # otherwise try a fuzzy match else: matching_worms_record = worms.aphia_record_by_fuzzy_taxon_name(original_scientific_name) if matching_worms_record is not None: worms_match_result = 'fuzzy’ # if either match succeeds extract the LSID for the taxon if matching_worms_record is not None: worms_lsid = matching_worms_record['lsid'] # @END find_matching_worms_record Finding matching WoRMS records in the data cleaning script YesWorkflow annotation marking start of workflow step. End of workflow step. Variables serving as inputs and outputs to workflow step. Calls to WoRMS service wrapper functions.
- 14. YW rendering of script Workflow steps each delimited by @BEGIN and @END annotations in script. Data flowing into and out of find_matching_worms_record workflow step. YesWorkflow infers connections between workflow steps and what data flows through them by matching @IN and @OUT annotations.
- 15. # @BEGIN compose_cleaned_record # @IN original_record # @IN worms_lsid # @IN updated_scientific_name # @IN original_scientific_name # @IN updated_authorship # @IN original_authorship # @OUT cleaned_record cleaned_record = original_record cleaned_record['LSID'] = worms_lsid cleaned_record['WoRMsMatchResult'] = worms_match_result if updated_scientific_name is not None: cleaned_record['scientificName'] = updated_scientific_name cleaned_record['originalScientificName'] = original_scientific_name if updated_authorship is not None: cleaned_record['scientificNameAuthorship'] = updated_authorship cleaned_record['originalAuthor'] = original_authorship # @END compose_cleaned_record The compose_cleaned_record step in the data cleaning script
- 16. A function for cleaning one record def curate_taxon_name_and_author(self, input_record): # look up record for input taxon name in WoRMS taxonomic database is_exact_match, aphia_record = ( self._worms.aphia_record_by_taxon_name(input_record['TaxonName'])) if aphia_record is not None: # save taxon name and author values from input record in new fields input_record['OriginalName'] = input_record['TaxonName'] input_record['OriginalAuthor'] = input_record['Author'] # replace taxon name and author fields in input record with values in aphia record input_record['TaxonName'] = aphia_record['scientificname'] input_record['Author'] = aphia_record['authority'] # add new fields input_record['WoRMsExactMatch'] = is_exact_match input_record['lsid'] = aphia_record['lsid’] else: input_record['OriginalName'] = None input_record['OriginalAuthor'] = None input_record['WoRMsExactMatch'] = None input_record['lsid'] = None return input_record Factoring out the core functionality of a script into a reusable function is a natural step in script evolution.
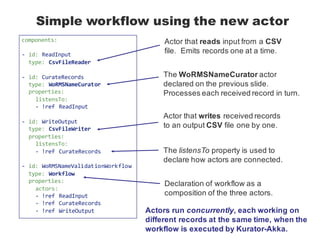
- 17. Declaring the function as an actor -‐ id: WoRMSNameCurator type: PythonClassActor properties: pythonClass: org.kurator.validation.actors.WoRMSCurator.WoRMSCurator onData: curate_taxon_name_and_author Actor type identifier referenced when composing a workflow that uses the actor. Python class declaring the function as a method (optional). Name of Python function called for each data item received by the actor at run time. Besides this block of YAML, no additional code needs to be written to convert the function into an actor.
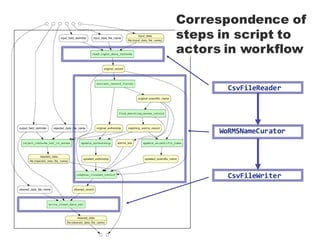
- 18. Simple workflow using the new actor components: -‐ id: ReadInput type: CsvFileReader -‐ id: CurateRecords type: WoRMSNameCurator properties: listensTo: -‐ !ref ReadInput -‐ id: WriteOutput type: CsvFileWriter properties: listensTo: -‐ !ref CurateRecords -‐ id: WoRMSNameValidationWorkflow type: Workflow properties: actors: -‐ !ref ReadInput -‐ !ref CurateRecords -‐ !ref WriteOutput Actor that reads input from a CSV file. Emits records one at a time. The WoRMSNameCurator actor declared on the previous slide. Processes each received record in turn. Actor that writes received records to an output CSV file one by one. The listensTo property is used to declare how actors are connected. Declaration of workflow as a composition of the three actors. Actors run concurrently, each working on different records at the same time, when the workflow is executed by Kurator-Akka.
- 19. CsvFileReader WoRMSNameCurator CsvFileWriter Correspondence of steps in script to actors in workflow
- 20. Running the workflow $ ka -‐f WoRMS_name_validation.yaml < WoRMS_name_validation_input.csv ID,TaxonName,Author,OriginalName,OriginalAuthor,WoRMsExactMatch,lsid 37929,Architectonica reevei,"(Hanley, 1862)",Architectonica reevi,,false,urn:lsid:marinespecies.org:taxname:588206 37932,Rapana rapiformis,"(Born, 1778)",Rapana rapiformis,"(Von Born, 1778)",true,urn:lsid:marinespecies.org:taxname:140415 180593,Buccinum donomani,"(Linnaeus, 1758)",,,, 179963,Codakia paytenorum,"(Iredale, 1937)",Codakia paytenorum,"Iredale, 1937",true,urn:lsid:marinespecies.org:taxname:215841 0,Rissoa venusta,"Garrett, 1873",Rissoa venusta,,true,urn:lsid:marinespecies.org:taxname:607233 62156,Rissoa venusta,"Garrett, 1873",Rissoa venusta,Phil.,true,urn:lsid:marinespecies.org:taxname:607233 $ Workflow can be run at the command line. Actors can be created from simple scripts, and workflows can be run like scripts. Simple YAML files wire everything together. Actors can read and write standard input and output like any script.
- 21. The road ahead The immediate future § YesWorkflow support for graphically rendering Kurator-Akka workflows. § Combining prospective and retrospective provenance from workflow declarations and YesWorkflow-annotated scripts used as actors. § Enhancements to YAML workflow declarations enabling Akka support for creating and managing multiple instances of each actor for higher throughput. § Exploration of advanced Akka features in FP-Akka framework, followed by generalization of these features in Kurator-Akka so that all users can benefit. Future possibilities § Support for additional actor scripting languages (e.g., R). § Actor function wrappers for other workflow and dataflow systems. § Graphical user interface for composing and running workflows. § Your suggestions?













![# @BEGIN find_matching_worms_record
# @IN original_scientific_name
# @OUT matching_worms_record
# @OUT worms_lsid
worms_match_result = None
worms_lsid = None
# first try exact match of the scientific name against WoRMS
matching_worms_record = worms.aphia_record_by_exact_taxon_name(original_scientific_name)
if matching_worms_record is not None:
worms_match_result = 'exact'
# otherwise try a fuzzy match
else:
matching_worms_record = worms.aphia_record_by_fuzzy_taxon_name(original_scientific_name)
if matching_worms_record is not None:
worms_match_result = 'fuzzy’
# if either match succeeds extract the LSID for the taxon
if matching_worms_record is not None:
worms_lsid = matching_worms_record['lsid']
# @END find_matching_worms_record
Finding matching WoRMS records
in the data cleaning script
YesWorkflow annotation
marking start of workflow step.
End of workflow step.
Variables serving as inputs and
outputs to workflow step.
Calls to WoRMS service
wrapper functions.](https://image.slidesharecdn.com/tdwg2015kuratorvalidationpresented-151006023858-lva1-app6892/85/Data-cleaning-with-the-Kurator-toolkit-Bridging-the-gap-between-conventional-scripting-and-high-performance-workflow-automation-13-320.jpg)

![# @BEGIN compose_cleaned_record
# @IN original_record
# @IN worms_lsid
# @IN updated_scientific_name
# @IN original_scientific_name
# @IN updated_authorship
# @IN original_authorship
# @OUT cleaned_record
cleaned_record = original_record
cleaned_record['LSID'] = worms_lsid
cleaned_record['WoRMsMatchResult'] = worms_match_result
if updated_scientific_name is not None:
cleaned_record['scientificName'] = updated_scientific_name
cleaned_record['originalScientificName'] = original_scientific_name
if updated_authorship is not None:
cleaned_record['scientificNameAuthorship'] = updated_authorship
cleaned_record['originalAuthor'] = original_authorship
# @END compose_cleaned_record
The compose_cleaned_record step in
the data cleaning script](https://image.slidesharecdn.com/tdwg2015kuratorvalidationpresented-151006023858-lva1-app6892/85/Data-cleaning-with-the-Kurator-toolkit-Bridging-the-gap-between-conventional-scripting-and-high-performance-workflow-automation-15-320.jpg)
![A function for cleaning one record
def curate_taxon_name_and_author(self, input_record):
# look up record for input taxon name in WoRMS taxonomic database
is_exact_match, aphia_record = (
self._worms.aphia_record_by_taxon_name(input_record['TaxonName']))
if aphia_record is not None:
# save taxon name and author values from input record in new fields
input_record['OriginalName'] = input_record['TaxonName']
input_record['OriginalAuthor'] = input_record['Author']
# replace taxon name and author fields in input record with values in aphia record
input_record['TaxonName'] = aphia_record['scientificname']
input_record['Author'] = aphia_record['authority']
# add new fields
input_record['WoRMsExactMatch'] = is_exact_match
input_record['lsid'] = aphia_record['lsid’]
else:
input_record['OriginalName'] = None
input_record['OriginalAuthor'] = None
input_record['WoRMsExactMatch'] = None
input_record['lsid'] = None
return input_record
Factoring out the core functionality
of a script into a reusable function
is a natural step in script evolution.](https://image.slidesharecdn.com/tdwg2015kuratorvalidationpresented-151006023858-lva1-app6892/85/Data-cleaning-with-the-Kurator-toolkit-Bridging-the-gap-between-conventional-scripting-and-high-performance-workflow-automation-16-320.jpg)




























![Accumulo Summit 2015: Alternatives to Apache Accumulo's Java API [API]](https://cdn.slidesharecdn.com/ss_thumbnails/as2015-elserjosh-150501223739-conversion-gate02-thumbnail.jpg?width=560&fit=bounds)
![Accumulo Summit 2015: Ambari and Accumulo: HDP 2.3 Upcoming Features [Sponsored]](https://cdn.slidesharecdn.com/ss_thumbnails/as2015-hwx-sponsored-talkrinaldibillie-150501220921-conversion-gate02-thumbnail.jpg?width=560&fit=bounds)








































