Deep Learning using Keras
- 1. Deep Learning using Keras ALY OSAMA DEEP LEARNING USING KERAS - ALY OSAMA 18/30/2017
- 2. About Me Graduated in 2016 from Faculty of Engineering, Ainshames University Currently, Research Software Development Engineer, Microsoft Research (ATLC) ◦ Speech Recognition Team “Arabic Models” ◦ Natural Language Processing Team “Virtual Bot” Part Time Teaching Assistant in Ainshames University DEEP LEARNING USING KERAS - ALY OSAMA 28/30/2017 alyosamah at gmail dot com
- 3. My Goal Today Introduce you to KerasIntroduce Not teach you all of KerasTeach But make sure that you can use KerasMake DEEP LEARNING USING KERAS - ALY OSAMA 38/30/2017
- 4. Agenda Overview Intro to Keras in 30 Seconds Case Study: Predict the effect of Genetic Variants Keras in depth Data Representation Keras Models and Layers Activations, Losses and Optimizers Learning rate scheduler Metrics and Performance evaluation strategies Regularizers Saving and loading Model visualization Callbacks Keras Cheat Sheet, Examples and Models Keras and Visualization Resources and References DEEP LEARNING USING KERAS - ALY OSAMA 48/30/2017
- 5. Overview DEEP LEARNING USING KERAS - ALY OSAMA 58/30/2017
- 6. Your Background Generally speaking, neural networks are nonlinear machine learning models. ◦ They can be used for supervised or unsupervised learning. Deep learning refers to training neural nets with multiple layers. ◦ They are more powerful but only if you have lots of data to train them on. Keras is used to create neural network models 8/30/2017 DEEP LEARNING USING KERAS - ALY OSAMA 6
- 7. Keras Overview • Neural Network library written in python • Design to be simple and straightforward • Built on top of different deep learning libraries such as Tensorflow, Theano and CNTK What is Keras? • Simple • Highly modular • Deep enough to build models Why Keras? DEEP LEARNING USING KERAS - ALY OSAMA 78/30/2017
- 8. Keras: Backends TensorFlow, Theano, CNTK (Microsoft) Can deploy in production via TensorFlow Serving 8
- 9. Keras is a trend 8/30/2017 DEEP LEARNING USING KERAS - ALY OSAMA 9
- 10. When to use Keras ? If you're a beginner and interested in quickly implementing your ideas • Python + Keras: Super fast implementation, good extensibility If you want to do fundamental research in Deep Learning • Python + Tensorflow or PyTorch: Excellent extensibility DEEP LEARNING USING KERAS - ALY OSAMA 108/30/2017
- 11. DEEP LEARNING USING KERAS - ALY OSAMA 118/30/2017
- 12. Intro to Keras DEEP LEARNING USING KERAS - ALY OSAMA 128/30/2017
- 13. General Design Prepare your input and output tensors 1 Create first layer to handle input layer 2 Create last layer to handle output targets 3 Build any model you like in between 4 DEEP LEARNING USING KERAS - ALY OSAMA 138/30/2017
- 14. Keras Steps DEEP LEARNING USING KERAS - ALY OSAMA 148/30/2017
- 15. Keras in 30 Seconds (1) from keras.models import Sequential model = Sequential() from keras.layers import Dense, Activation model.add(Dense(units=64, input_dim=100)) model.add(Activation('relu')) model.add(Dense(units=10)) model.add(Activation('softmax')) 1. Sequential model ( a linear stack of layers ) DEEP LEARNING USING KERAS - ALY OSAMA 158/30/2017
- 16. Keras in 30 Seconds (2) model.compile(loss='categorical_crossentropy', optimizer='sgd', metrics=['accuracy']) model.compile(loss=keras.losses.categorical_crossentropy, optimizer=keras.optimizers.SGD(lr=0.01, momentum=0.9)) 2. Compile Model Also, you can further configure your optimizer DEEP LEARNING USING KERAS - ALY OSAMA 168/30/2017
- 17. Keras in 30 Seconds (3) model.fit(x_train, y_train, epochs=5, batch_size=32) model.train_on_batch(x_batch, y_batch) 3. Training You can feed data batches manualy loss_and_metrics = model.evaluate(x_test, y_test, batch_size=128) 4. Evaluation classes = model.predict(x_test, batch_size=128) 5. Prediction DEEP LEARNING USING KERAS - ALY OSAMA 178/30/2017
- 18. Case Study: Predict the effect of Genetic Variants DEEP LEARNING USING KERAS - ALY OSAMA 188/30/2017
- 19. Problem Currently this interpretation of genetic mutations is being done manually. This is a very time- consuming task where a clinical pathologist has to manually review and classify every single genetic mutation based on evidence from text-based clinical literature. The Challenge is to develop classification models which analyze abstracts of medical articles and, based on their content accurately determine a mutation effect (9 classes) of the genes discussed in them DEEP LEARNING USING KERAS - ALY OSAMA 198/30/2017
- 20. Input dataset ( Variations + Text ) 8/30/2017 DEEP LEARNING USING KERAS - ALY OSAMA 20
- 21. Text (20%) – Wow very Huge 0||Cyclin-dependent kinases (CDKs) regulate a variety of fundamental cellular processes. CDK10 stands out as one of the last orphan CDKs for which no activating cyclin has been identified and no kinase activity revealed. Previous work has shown that CDK10 silencing increases ETS2 (v-ets erythroblastosis virus E26 oncogene homolog 2)-driven activation of the MAPK pathway, which confers tamoxifen resistance to breast cancer cells. The precise mechanisms by which CDK10 modulates ETS2 activity, and more generally the functions of CDK10, remain elusive. Here we demonstrate that CDK10 is a cyclin-dependent kinase by identifying cyclin M as an activating cyclin. Cyclin M, an orphan cyclin, is the product of FAM58A, whose mutations cause STAR syndrome, a human developmental anomaly whose features include toe syndactyly, telecanthus, and anogenital and renal malformations. We show that STAR syndrome-associated cyclin M mutants are unable to interact with CDK10. Cyclin M silencing phenocopies CDK10 silencing in increasing c-Raf and in conferring tamoxifen resistance to breast cancer cells. CDK10/cyclin M phosphorylates ETS2 in vitro, and in cells it positively controls ETS2 degradation by the proteasome. ETS2 protein levels are increased in cells derived from a STAR patient, and this increase is attributable to decreased cyclin M levels. Altogether, our results reveal an additional regulatory mechanism for ETS2, which plays key roles in cancer and development. They also shed light on the molecular mechanisms underlying STAR syndrome.Cyclin-dependent kinases (CDKs) play a pivotal role in the control of a number of fundamental cellular processes (1). The human genome contains 21 genes encoding proteins that can be considered as members of the CDK family owing to their sequence similarity with bona fide CDKs, those known to be activated by cyclins (2). Although discovered almost 20 y ago (3, 4), CDK10 remains one of the two CDKs without an identified cyclin partner. This knowledge gap has largely impeded the exploration of its biological functions. CDK10 can act as a positive cell cycle regulator in some cells (5, 6) or as a tumor suppressor in others (7, 8). CDK10 interacts with the ETS2 (v-ets erythroblastosis virus E26 oncogene homolog 2) transcription factor and inhibits its transcriptional activity through an unknown mechanism (9). CDK10 knockdown derepresses ETS2, which increases the expression of the c-Raf protein kinase, activates the MAPK pathway, and induces resistance of MCF7 cells to tamoxifen (6).Here, we deorphanize CDK10 by identifying cyclin M, the product of FAM58A, as a binding partner. Mutations in this gene that predict absence or truncation of cyclin M are associated with STAR syndrome, whose features include toe syndactyly, telecanthus, and anogenital and renal malformations in heterozygous females (10). However, both the functions of cyclin M and the pathogenesis of STAR syndrome remain unknown. We show that a recombinant CDK10/cyclin M heterodimer is an active protein kinase that phosphorylates ETS2 in vitro. Cyclin M silencing phenocopies CDK10 silencing in increasing c-Raf and phospho-ERK expression levels and in inducing tamoxifen resistance in estrogen receptor (ER)+ breast cancer cells. We show that CDK10/cyclin M positively controls ETS2 degradation by the proteasome, through the phosphorylation of two neighboring serines. Finally, we detect an increased ETS2 expression level in cells derived from a STAR patient, and we demonstrate that it is attributable to the decreased cyclin M expression level observed in these cells.Previous SectionNext SectionResultsA yeast two-hybrid (Y2H) screen unveiled an interaction signal between CDK10 and a mouse protein whose C-terminal half presents a strong sequence homology with the human FAM58A gene product [whose proposed name is cyclin M (11)]. We thus performed Y2H mating assays to determine whether human CDK10 interacts with human cyclin M (Fig. 1 A–C). The longest CDK10 isoform (P1) expressed as a bait protein produced a strong interaction phenotype with full-length cyclin M (expressed as a prey protein) but no detectable phenotype with cyclin D1, p21 (CIP1), and Cdi1 (KAP), which are known binding partners of other CDKs (Fig. 1B). CDK1 and CDK3 also produced Y2H signals with cyclin M, albeit notably weaker than that observed with CDK10 (Fig. 1B). An interaction phenotype was also observed between full-length cyclin M and CDK10 proteins expressed as bait and prey, respectively (Fig. S1A). We then tested different isoforms of CDK10 and cyclin M originating from alternative gene splicing, and two truncated cyclin M proteins corresponding to the hypothetical products of two mutated FAM58A genes found in STAR syndrome patients (10). None of these shorter isoforms produced interaction phenotypes (Fig. 1 A and C and Fig. S1A).Fig. 1.In a new window Download PPTFig. 1.CDK10 and cyclin M form an interaction complex. (A) Schematic representation of the different protein isoforms analyzed by Y2H assays. Amino acid numbers are indicated. Black boxes indicate internal deletions. The red box indicates a differing amino acid sequence compared with CDK10 P1. (B) Y2H assay between a set of CDK proteins expressed as baits (in fusion to the LexA DNA binding domain) and CDK interacting proteins expressed as preys (in fusion to the B42 transcriptional activator). pEG202 and pJG4-5 are the empty bait and prey plasmids expressing LexA and B42, respectively. lacZ was used as a reporter gene, and blue yeast are indicative of a Y2H interaction phenotype. (C) Y2H assay between the different CDK10 and cyclin M isoforms. The amino-terminal region of ETS2, known to interact with CDK10 (9), was also assayed. (D) Western blot analysis of Myc-CDK10 (wt or kd) and CycM-V5-6His expression levels in transfected HEK293 cells. (E) Western blot analysis of Myc-CDK10 (wt or kd) immunoprecipitates obtained using the anti-Myc antibody. “Inputs” correspond to 10 μg total lysates obtained from HEK293 cells coexpressing Myc-CDK10 (wt or kd) and CycM-V5-6His. (F) Western blot analysis of immunoprecipitates obtained using the anti-CDK10 antibody or a control goat antibody, from human breast cancer MCF7 cells. “Input” corresponds to 30 μg MCF7 total cell lysates. The lower band of the doublet observed on the upper panel comigrates with the exogenously expressed untagged CDK10 and thus corresponds to endogenous CDK10. The upper band of the doublet corresponds to a nonspecific signal, as demonstrated by it insensitivity to either overexpression of CDK10 (as seen on the left lane) or silencing of CDK10 (Fig. S2B). Another experiment with a longer gel migration is shown in Fig. S1D.Next we examined the ability of CDK10 and cyclin M to interact when expressed in human cells (Fig. 1 D and E). We tested wild-type CDK10 (wt) and a kinase dead (kd) mutant bearing a D181A amino acid substitution that abolishes ATP binding (12). We expressed cyclin M-V5-6His and/or Myc-CDK10 (wt or kd) in a human embryonic kidney cell line (HEK293). The expression level of cyclin M-V5-6His was significantly increased upon coexpression with Myc-CDK10 (wt or kd) and, to a lesser extent, that of Myc-CDK10 (wt or kd) was increased upon coexpression with cyclin M-V5-6His (Fig. 1D). We then immunoprecipitated Myc-CDK10 proteins and detected the presence of cyclin M in the CDK10 (wt) and (kd) immunoprecipitates only when these proteins were coexpressed pair-wise (Fig. 1E). We confirmed these observations by detecting the presence of Myc-CDK10 in cyclin M-V5-6His immunoprecipitates (Fig. S1B). These experiments confirmed the lack of robust interaction between the CDK10.P2 isoform and cyclin M (Fig. S1C). To detect the interaction between endogenous proteins, we performed immunoprecipitations on nontransfected MCF7 cells derived from a human breast cancer. CDK10 and cyclin M antibodies detected their cognate endogenous proteins by Western blotting. We readily detected cyclin M in immunoprecipitates obtained with the CDK10 antibody but not with a control antibody (Fig. 1F). These results confirm the physical interaction between CDK10 and cyclin M in human cells.To unveil a hypothesized CDK10/cyclin M protein kinase activity, we produced GST-CDK10 and StrepII-cyclin M fusion proteins in insect cells, either individually or in combination. We observed that GST-CDK10 and StrepII-cyclin M copurified, thus confirming their interaction in yet another cellular model (Fig. 2A). We then performed in vitro kinase assays with purified proteins, using histone H1 as a generic substrate. Histone H1 phosphorylation was detected only from lysates of cells coexpressing GST-CDK10 and StrepII-cyclin M. No phosphorylation was detected when GST- CDK10 or StrepII-cyclin M were expressed alone, or when StrepII-cyclin M was coexpressed with GST-CDK10(kd) (Fig. 2A). Next we investigated whether ETS2, which is known to interact with CDK10 (9) (Fig. 1C), is a phosphorylation substrate of CDK10/cyclin M. We detected strong phosphorylation of ETS2 by the GST- CDK10/StrepII-cyclin M purified heterodimer, whereas no phosphorylation was detected using GST-CDK10 alone or GST-CDK10(kd)/StrepII-cyclin M heterodimer (Fig. 2B).Fig. 2.In a new window Download PPTFig. 2.CDK10 is a cyclin M-dependent protein kinase. (A) In vitro protein kinase assay on histone H1. Lysates from insect cells expressing different proteins were purified on a glutathione Sepharose matrix to capture GST-CDK10(wt or kd) fusion proteins alone, or in complex with STR-CycM fusion protein. Purified protein expression levels were analyzed by Western blots (Top and Upper Middle). The kinase activity was determined by autoradiography of histone H1, whose added amounts were visualized by Coomassie staining (Lower Middle and Bottom). (B) Same as in A, using purified recombinant 6His-ETS2 as a substrate.CDK10 silencing has been shown to increase ETS2-driven c-RAF transcription and to activate the MAPK pathway (6). We investigated whether cyclin M is also involved in this regulatory pathway. To aim at a highly specific silencing, we used siRNA pools (mix of four different siRNAs) at low final concentration (10 nM). Both CDK10 and cyclin M siRNA pools silenced the expression of their cognate targets (Fig. 3 A and C and Fig. S2) and, interestingly, the cyclin M siRNA pool also caused a marked decrease in CDK10 protein level (Fig. 3A and Fig. S2B). These results, and those shown in Fig. 1D, suggest that cyclin M binding stabilizes CDK10. Cyclin M silencing induced an increase in c-Raf protein and mRNA levels (Fig. 3 B and C) and in phosphorylated ERK1 and ERK2 protein levels (Fig. S3B), similarly to CDK10 silencing. As expected from these effects (6), CDK10 and cyclin M silencing both decreased the sensitivity of ER+ MCF7 cells to tamoxifen, to a similar extent. The combined silencing of both genes did not result in a higher resistance to the drug (Fig. S3C). Altogether, these observations demonstrate a functional interaction between cyclin M and CDK10, which negatively controls ETS2.Fig. 3.In a new window Download PPTFig. 3.Cyclin M silencing up-regulates c-Raf expression. (A) Western blot analysis of endogenous CDK10 and cyclin M expression levels in MCF7 cells, in response to siRNA-mediated gene silencing. (B) Western blot analysis of endogenous c-Raf expression levels in MCF7 cells, in response to CDK10 or cyclin M silencing. A quantification is shown in Fig. S3A. (C) Quantitative RT-PCR analysis of CDK10, cyclin M, and c-Raf mRNA levels, in response to CDK10 (Upper) or cyclin M (Lower) silencing. **P ≤ 0.01; ***P ≤ 0.001.We then wished to explore the mechanism by which CDK10/cyclin M controls ETS2. ETS2 is a short-lived protein degraded by the proteasome (13). A straightforward hypothesis is that CDK10/cyclin M positively controls ETS2 degradation. We thus examined the impact of CDK10 or cyclin M silencing on ETS2 expression levels. The silencing of CDK10 and that of cyclin M caused an increase in the expression levels of an exogenously expressed Flag-ETS2 protein (Fig. S4A), as well as of the endogenous ETS2 protein (Fig. 4A). This increase is not attributable to increased ETS2 mRNA levels, which marginally fluctuated in response to CDK10 or cyclin M silencing (Fig. S4B). We then examined the expression levels of the Flag-tagged ETS2 protein when expressed alone or in combination with Myc-CDK10 or -CDK10(kd), with or without cyclin M-V5-6His. Flag-ETS2 was readily detected when expressed alone or, to a lesser extent, when coexpressed with CDK10(kd). However, its expression level was dramatically decreased when coexpressed with CDK10 alone, or with CDK10 and cyclin M (Fig. 4B). These observations suggest that endogenous cyclin M levels are in excess compared with those of CDK10 in MCF7 cells, and they show that the major decrease in ETS2 levels observed upon CDK10 coexpression involves CDK10 kinase activity. Treatment of cells coexpressing Flag-ETS2, CDK10, and cyclin M with the proteasome inhibitor MG132 largely rescued Flag-ETS2 expression levels (Fig. 4B).Fig. 4.In a new window Download PPTFig. 4.CDK10/cyclin M controls ETS2 stability in human cancer derived cells. (A) Western blot analysis of endogenous ETS2 expression levels in MCF7 cells, in response to siRNA-mediated CDK10 and/or cyclin M silencing. A quantification is shown in Fig. S4B. (B) Western blot analysis of exogenously expressed Flag-ETS2 protein levels in MCF7 cells cotransfected with empty vectors or coexpressing Myc-CDK10 (wt or kd), or Myc-CDK10/CycM-V5-6His. The latter cells were treated for 16 h with the MG132 proteasome inhibitor. Proper expression of CDK10 and cyclin M tagged proteins was verified by Western blot analysis. (C and D) Western blot analysis of expression levels of exogenously expressed Flag-ETS2 wild-type or mutant proteins in MCF7 cells, in the absence of (C) or in response to (D) Myc-CDK10/CycM-V5-6His expression. Quantifications are shown in Fig. S4 C and D.A mass spectrometry analysis of recombinant ETS2 phosphorylated by CDK10/cyclin M in vitro revealed the existence of multiple phosphorylated residues, among which are two neighboring phospho-serines (at positions 220 and 225) that may form a phosphodegron (14) (Figs. S5–S8). To confirm this finding, we compared the phosphorylation level of recombinant ETS2wt with that of ETS2SASA protein, a mutant bearing alanine substitutions of these two serines. As expected from the existence of multiple phosphorylation sites, we detected a small but reproducible, significant decrease of phosphorylation level of ETS2SASA compared with ETS2wt (Fig. S9), thus confirming that Ser220/Ser225 are phosphorylated by CDK10/cyclin M. To establish a direct link between ETS2 phosphorylation by CDK10/cyclin M and degradation, we examined the expression levels of Flag-ETS2SASA. In the absence of CDK10/cyclin M coexpression, it did not differ significantly from that of Flag-ETS2. This is contrary to that of Flag-ETS2DBM, bearing a deletion of the N- terminal destruction (D-) box that was previously shown to be involved in APC-Cdh1–mediated degradation of ETS2 (13) (Fig. 4C). However, contrary to Flag-ETS2 wild type, the expression level of Flag-ETS2SASA remained insensitive to CDK10/cyclin M coexpression (Fig. 4D). Altogether, these results suggest that CDK10/cyclin M directly controls ETS2 degradation through the phosphorylation of these two serines.Finally, we studied a lymphoblastoid cell line derived from a patient with STAR syndrome, bearing FAM58A mutation c.555+1G>A, predicted to result in aberrant splicing (10). In ………………. 8/30/2017 DEEP LEARNING USING KERAS - ALY OSAMA 21
- 22. Frequency of Classes 8/30/2017 DEEP LEARNING USING KERAS - ALY OSAMA 22
- 23. Gene and Variation Statistics 8/30/2017 DEEP LEARNING USING KERAS - ALY OSAMA 23
- 24. More Statistics 8/30/2017 DEEP LEARNING USING KERAS - ALY OSAMA 24
- 25. For more Statistics https://www.kaggle.com/headsortails/ personalised-medicine-eda-with-tidy-r 8/30/2017DEEP LEARNING USING KERAS - ALY OSAMA 25
- 26. Baseline Solution Clean Text Data Train Doc2Vec with Text SVD for one hot encoding for Genes and Variants Get Features ( Embedding(300) + SVD(25) + SVD(25) ) = 350 Train Keras Model Prediction DEEP LEARNING USING KERAS - ALY OSAMA 268/30/2017
- 27. Baseline Solution 8/30/2017 DEEP LEARNING USING KERAS - ALY OSAMA 27 Code: https://www.kaggle.com/alyosama/doc2vec-with-keras-0-77/ Text Gene Variation Text Cleaning Train Doc2Vec(300) One Hot Encoding One Hot Encoding SVD (25 dim) SVD (25 dim) Input Vector (350) Class One Hot Encoding Output Vector (9)
- 28. Keras in depth DEEP LEARNING USING KERAS - ALY OSAMA 288/30/2017
- 29. 1. Data Representation Discrete Variables ◦ Numeric variables that have a countable number of values between any two values. E.g., the number of children in a family - 3. Continuous Variables ◦ Numeric variables that have an infinite number of values between any two values. E.g., temperature - 25.9 Categorical variables ◦ A finite number of categories or distinct groups. Categorical data might not have a logical order. E.g., gender - male/female DEEP LEARNING USING KERAS - ALY OSAMA 298/30/2017
- 30. 1. Data Representation - Utile Although you can use any other method for feature preprocessing, keras has a couple of utilities to help, such as: ◦ To_categorical (to one-hot encode data) ◦ Text preprocessing utilities, such as tokenizing ◦ LabelEncoder “ using Skikit Learn” 8/30/2017 DEEP LEARNING USING KERAS - ALY OSAMA 30 from sklearn.preprocessing import LabelEncoder from keras.utils import np_utils encoder = LabelEncoder() encoder.fit(Y) encoded_Y = encoder.transform(Y) # convert integers to dummy variables (i.e. one hot encoded) dummy_y = np_utils.to_categorical(encoded_Y)
- 31. 2. Keras Models - Sequential Models 1. Sequential Models : stack of layers data_dim = 16 timesteps = 8 model = Sequential() model.add(LSTM(32, return_sequences=True, input_shape=(timesteps, data_dim))) model.add(LSTM(32, return_sequences=True)) model.add(LSTM(32)) model.add(Dense(10, activation='softmax')) model.compile(loss='categorical_crossentropy ', optimizer='rmsprop', metrics=['accuracy']) DEEP LEARNING USING KERAS - ALY OSAMA 318/30/2017
- 32. 2. Keras Models - Model Class API 2. Model Class API from keras.models import Model from keras.layers import Input, Dense a = Input(shape=(32,)) b = Dense(32)(a) model = Model(inputs=a, outputs=b) • Optimized over all outputs Graph model • allows for two or more independent networks to diverge or merge • Allows for multiple separate inputs or outputs • Different merging layers (sum or concatenate) DEEP LEARNING USING KERAS - ALY OSAMA 328/30/2017
- 33. 3. Keras Layers Layers are used to define what your architecture looks like Examples of layers are: ◦ Dense layers (this is the normal, fully-connected layer) ◦ Convolutional layers (applies convolution operations on the previous layer) ◦ Pooling layers (used after convolutional layers) ◦ Dropout layers (these are used for regularization, to avoid overfitting) 8/30/2017 DEEP LEARNING USING KERAS - ALY OSAMA 33
- 34. 3.1 Keras Layers Keras has a number of pre-built layers. Notable examples include: Regular dense, MLP type Recurrent layers, LSTM, GRU, etc keras.layers.core.Dense(units, activation=None, use_bias=True, kernel_initializer='glorot_uniform', bias_initializer='zeros', kernel_regularizer=None, bias_regularizer=None, activity_regularizer=None, kernel_constraint=None, bias_constraint=None) keras.layers.recurrent.LSTM(units, activation='tanh', recurrent_activation='hard_sigmoid', use_bias=True, kernel_initializer='glorot_uniform', recurrent_initializer='orthogonal', bias_initializer='zeros', unit_forget_bias=True, kernel_regularizer=None, recurrent_regularizer=None, bias_regularizer=None, activity_regularizer=None, kernel_constraint=None, recurrent_constraint=None, bias_constraint=None, dropout=0.0, recurrent_dropout=0.0) DEEP LEARNING USING KERAS - ALY OSAMA 348/30/2017
- 35. 3.2 Keras Layers 2D Convolutional layers Autoencoders can be built with any other type of layer keras.layers.convolutional.Conv2D(filters, kernel_size, strides=(1, 1), padding='valid', data_format=None, dilation_rate=(1, 1), activation=None, use_bias=True, kernel_initializer='glorot_uniform', bias_initializer='zeros', kernel_regularizer=None, bias_regularizer=None, activity_regularizer=None, kernel_constraint=None, bias_constraint=None) from keras.layers import Dense, Activation model.add(Dense(units=32, input_dim=512)) model.add(Activation('relu')) model.add(Dense(units=512)) model.add(Activation(‘sigmoid')) DEEP LEARNING USING KERAS - ALY OSAMA 358/30/2017
- 36. 3.3 Keras Layers Other types of layer include: Noise Pooling Normalization Embedding And many more... DEEP LEARNING USING KERAS - ALY OSAMA 368/30/2017
- 37. 4. Activations All your favorite activations are available: 1. Sigmoid, tanh, ReLu, softplus, hard sigmoid, linear 2. Advanced activations implemented as a layer (after desired neural layer) 3. Advanced activations: LeakyReLu, PReLu, ELU, Parametric Softplus, Thresholded linear and Thresholded Relu DEEP LEARNING USING KERAS - ALY OSAMA 378/30/2017
- 38. 5. Losses (Objectives) binary_crossentropy categorical_crossentropy mean_squared_error sparse_categorical_crossentropy kullback_leibler_divergence poisson cosine_proximity mean_absolute_error mean_absolute_percentage_error mean_squared_logarithmic_error squared_hinge hinge categorical_hinge logcosh from keras.utils.np_utils import to_categorical categorical_labels = to_categorical(int_labels, num_classes=None) model.compile(loss='mean_squared_error', optimizer='sgd') DEEP LEARNING USING KERAS - ALY OSAMA 388/30/2017 In case you want to convert your categorical data:
- 39. 6. Optimizers An optimizer is one of the two arguments required for compiling a Keras model: from keras import optimizers model = Sequential() model.add(Dense(64, kernel_initializer='uniform', input_shape=(10,))) model.add(Activation('tanh’)) sgd = optimizers.SGD(lr=0.01, decay=1e-6, momentum=0.9, nesterov=True) model.compile(loss='mean_squared_error', optimizer=sgd) or you can call it by its name. In the latter case, the default parameters for the optimizer will be used. # pass optimizer by name: default parameters will be used model.compile(loss='mean_squared_error', optimizer='sgd') DEEP LEARNING USING KERAS - ALY OSAMA 398/30/2017
- 40. 6. Optimizers - More 1. SGD :Stochastic gradient descent optimizer. 2. RMSprop: RMSProp optimizer is usually a good choice for recurrent neural networks. Adagrad 3. Nadam : Much like Adam is essentially RMSprop with momentum, Nadam is Adam RMSprop with Nesterov momentum 4. Adadelta 5. Adam You can also use a wrapper class for native TensorFlow optimizers TFOptimizer:. keras.optimizers.SGD(lr=0.01, momentum=0.0, decay=0.0, nesterov=False) keras.optimizers.RMSprop(lr=0.001, rho=0.9, epsilon=1e-08, decay=0.0) keras.optimizers.Adagrad(lr=0.01, epsilon=1e-08, decay=0.0) keras.optimizers.Adadelta(lr=1.0, rho=0.95, epsilon=1e-08, decay=0.0) keras.optimizers.Adam(lr=0.001, beta_1=0.9, beta_2=0.999, epsilon=1e-08, decay=0.0) keras.optimizers.Nadam(lr=0.002, beta_1=0.9, beta_2=0.999, epsilon=1e-08, schedule_decay=0.004) keras.optimizers.TFOptimizer(optimizer) DEEP LEARNING USING KERAS - ALY OSAMA 408/30/2017
- 41. 7. Learning Rate Scheduler In Keras you have two types of learning rate schedule: a time-based learning rate schedule. a drop-based learning rate schedule. DEEP LEARNING USING KERAS - ALY OSAMA 418/30/2017
- 42. 7.1 Time Based Learning Rate if we use the initial learning rate value of 0.1 and the decay of 0.001, the first 5 epochs will adapt the learning rate as follows: Epoch Learning Rate 1 0.1 2 0.0999000999 3 0.0997006985 4 0.09940249103 5 0.09900646517 LearningRate = LearningRate * 1/(1 + decay * epoch) DEEP LEARNING USING KERAS - ALY OSAMA 428/30/2017
- 43. 7.2 Drop-Based Learning Rate LearningRateScheduler: a function that takes an epoch index as input (integer, indexed from 0) and returns a new learning rate as output (float). # learning rate schedule def step_decay(epoch): initial_lrate = 0.1 drop = 0.5 epochs_drop = 10.0 lrate = initial_lrate * math.pow(drop, math.floor((1+epoch)/epochs_drop)) return lrate lrate = LearningRateScheduler(step_decay) callbacks_list = [lrate] # Fit the model model.fit(X, Y, validation_split=0.33, epochs=50, batch_size=28, callbacks=callbacks_list, verbose=2) DEEP LEARNING USING KERAS - ALY OSAMA 438/30/2017
- 44. 7.3 Tips for Using Learning Rate Increase the initial learning rate. • Because the learning rate will very likely decrease, start with a larger value to decrease from. A larger learning rate will result in a lot larger changes to the weights, at least in the beginning, allowing you to benefit from the fine tuning later. Use a large momentum. • Using a larger momentum value will help the optimization algorithm to continue to make updates in the right direction when your learning rate shrinks to small values. Experiment with different schedules. • It will not be clear which learning rate schedule to use so try a few with different configuration options and see what works best on your problem. Also try schedules that change exponentially and even schedules that respond to the accuracy of your model on the training or test datasets. DEEP LEARNING USING KERAS - ALY OSAMA 448/30/2017
- 45. 8. Metrics 1. Accuracy • binary_accuracy • categorical_accuracy • sparse_categorical_accuracy • top_k_categorical_accuracy • sparse_top_k_categorical_accuracy 2. Precision 3. Recall 4. FScore from keras import metrics model.compile(loss='categorical_crossentropy', optimizer='adadelta’, metrics=['accuracy', 'f1score', 'precision', 'recall']) 1280/5640 [=====>........................] - ETA: 20s - loss: 1.5566 - fmeasure: 0.8134 - precision: 0.8421 - recall: 0.7867 DEEP LEARNING USING KERAS - ALY OSAMA 458/30/2017
- 46. 8. Metrics - Custom Custom metrics can be passed at the compilation step. The function would need to take (y_true, y_pred) as arguments and return a single tensor value. import keras.backend as K def mean_pred(y_true, y_pred): return K.mean(y_pred) model.compile(optimizer='rmsprop', loss='binary_crossentropy', metrics=['accuracy', mean_pred]) DEEP LEARNING USING KERAS - ALY OSAMA 468/30/2017
- 47. 9. Performance evaluation strategies The large amount of data and the complexity of the models require very long training times. Keras provides a three convenient ways of evaluating your deep learning algorithms : ◦ Use an automatic verification dataset. ◦ Use a manual verification dataset. ◦ Use a manual k-Fold Cross Validation. DEEP LEARNING USING KERAS - ALY OSAMA 478/30/2017
- 48. 9.1 Automatic Verification Dataset Keras can separate a portion of your training data into a validation dataset and evaluate the performance of your model on that validation dataset each epoch model.fit(X, Y, validation_split=0.33, epochs=150, batch_size=10) ... Epoch 148/150 514/514 [==============================] - 0s - loss: 0.5219 - acc: 0.7354 - val_loss: 0.5414 - val_acc: 0.7520 Epoch 149/150 514/514 [==============================] - 0s - loss: 0.5089 - acc: 0.7432 - val_loss: 0.5417 - val_acc: 0.7520 Epoch 150/150 514/514 [==============================] - 0s - loss: 0.5148 - acc: 0.7490 - val_loss: 0.5549 - val_acc: 0.7520 DEEP LEARNING USING KERAS - ALY OSAMA 488/30/2017
- 49. 9.2 Manual Verification Dataset Keras also allows you to manually specify the dataset to use for validation during training. # MLP with manual validation set from sklearn.model_selection import train_test_split import numpy # fix random seed for reproducibility seed = 7 numpy.random.seed(seed) X_train, X_test, y_train, y_test = train_test_split(X, Y, test_size=0.33, random_state=seed) # Fit the model model.fit(X_train, y_train, validation_data=(X_test,y_test), epochs=150, batch_size=10) DEEP LEARNING USING KERAS - ALY OSAMA 498/30/2017
- 50. 9.3 Manual k-Fold Cross Validation from sklearn.model_selection import StratifiedKFold kfold = StratifiedKFold(n_splits=10, shuffle=True, random_state=seed) cvscores = [] for train, test in kfold.split(X, Y): # create model model = Sequential() model.add(Dense(12, input_dim=8, activation='relu')) model.add(Dense(8, activation='relu')) model.add(Dense(1, activation='sigmoid')) model.compile(loss='binary_crossentropy', optimizer='adam', metrics=['accuracy']) # Fit the model model.fit(X[train], Y[train], epochs=150, batch_size=10, verbose=0) # evaluate the model scores = model.evaluate(X[test], Y[test], verbose=0) print("%s: %.2f%%" % (model.metrics_names[1], scores[1]*100)) cvscores.append(scores[1] * 100) print("%.2f%% (+/- %.2f%%)" % (numpy.mean(cvscores), numpy.std(cvscores))) acc: 77.92% acc: 68.83% acc: 72.73% acc: 64.94% acc: 77.92% acc: 35.06% acc: 74.03% acc: 68.83% acc: 34.21% acc: 72.37% 64.68% (+/- 15.50%) DEEP LEARNING USING KERAS - ALY OSAMA 508/30/2017
- 51. 10. Regularizers Nearly everything in Keras can be regularized to avoid overfitting In addition to the Dropout layer, there are all sorts of other regularizers available, such as: ◦ Weight regularizers ◦ Bias regularizers ◦ Activity regularizers 8/30/2017 DEEP LEARNING USING KERAS - ALY OSAMA 51 from keras import regularizers model.add(Dense(64, input_dim=64, kernel_regularizer=regularizers.l2(0.01), activity_regularizer=regularizers.l1(0.01)))
- 52. 11. Architecture/Weight Saving and Loading Saving/loading whole models (architecture + weights + optimizer state) You can also save/load only a model's architecture or weights only. from keras.models import load_model model.save('my_model.h5') # creates a HDF5 file 'my_model.h5’ del model # deletes the existing model # returns a compiled model # identical to the previous one model = load_model('my_model.h5') DEEP LEARNING USING KERAS - ALY OSAMA 528/30/2017
- 53. 12. Model Visualization from keras.utils import plot_model plot_model(model, to_file='model.png') In case you want an image of your model : DEEP LEARNING USING KERAS - ALY OSAMA 538/30/2017
- 54. 13. Callbacks Allow for function call during training ◦ Callbacks can be called at different points of training (batch or epoch) ◦ Existing callbacks: Early Stopping, weight saving after epoch ◦ Easy to build and implement, called in training function, fit() DEEP LEARNING USING KERAS - ALY OSAMA 548/30/2017
- 55. 13.1 Callbacks - Examples TerminateOnNaN : Callback that terminates training when a NaN loss is encountered. EarlyStopping: Stop training when a monitored quantity has stopped improving. ModelCheckpoint : Save the model after every epoch. ReduceLROnPlateau: Reduce learning rate when a metric has stopped improving. keras.callbacks.ModelCheckpoint(filepath, monitor='val_loss', verbose=0, save_best_only=False, save_weights_only=False, mode='auto', period=1) keras.callbacks.ReduceLROnPlateau(monitor='val_loss', factor=0.1, patience=10, verbose=0, mode='auto', epsilon=0.0001, cooldown=0, min_lr=0) Also, You can create a custom callback by extending the base class keras.callbacks.Callback DEEP LEARNING USING KERAS - ALY OSAMA 558/30/2017
- 56. 13.2 Keras + TensorBoard: Visualizing Learning tbCallBack= keras.callbacks.TensorBoard(log_dir='./logs', histogram_freq=0, batch_size=32, write_graph=True, write_grads=False, write_images=False, embeddings_freq=0, embeddings_layer_names=None, embeddings_metadata=None) • TensorBoard is a visualization tool provided with TensorFlow. • This callback writes a log for TensorBoard, which allows you to visualize dynamic graphs of your training and test metrics, as well as activation histograms for the different layers in your model. # Use your terminal tensorboard --logdir path_to_current_dir/logs DEEP LEARNING USING KERAS - ALY OSAMA 568/30/2017
- 57. 14. Keras + Scikit You can integrate Keras models into a Scikit-learn Pipeline ◦ There are special wrapper functions available on Keras to help you implement the methods that are expected by a scikit-learn classifier, such as fit(), predict(), predict_proba(),etc. ◦ You can also use things like scikit-learn’s grid_search, to do model selection on Keras models, to decide what are the best hyperparameters for a given task. 8/30/2017 DEEP LEARNING USING KERAS - ALY OSAMA 57
- 58. Keras Cheat Sheet and Examples DEEP LEARNING USING KERAS - ALY OSAMA 588/30/2017
- 59. Model Architecture - Cheat Sheet 8/30/2017 DEEP LEARNING USING KERAS - ALY OSAMA 59
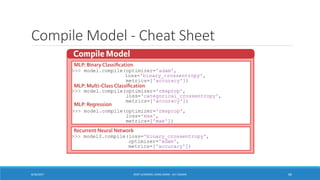
- 60. Compile Model - Cheat Sheet 8/30/2017 DEEP LEARNING USING KERAS - ALY OSAMA 60
- 61. Training, Evaluating and Prediction – Cheat Sheet 8/30/2017 DEEP LEARNING USING KERAS - ALY OSAMA 61
- 65. Check this out You can implement a difficult problem using Keras in 10 lines of code 65
- 66. Model Code 66
- 67. Keras + Visualization DEEP LEARNING USING KERAS - ALY OSAMA 678/30/2017
- 68. Visualization Plugin: Toolbox for Keras Dense layer visualization: How can we assess whether a network is over/under fitting or generalizing well? 8/30/2017 DEEP LEARNING USING KERAS - ALY OSAMA 68 Conv filter visualization Attention Maps :How can we assess whether a network is attending to correct parts of the image in order to generate a decision? https://github.com/raghakot/keras-vis
- 69. More Visualization tools: T-SNE t-Distributed Stochastic Neighbor Embedding (t-SNE) is a (prize- winning) technique for dimensionality reduction that is particularly well suited for the visualization of high-dimensional datasets. DEEP LEARNING USING KERAS - ALY OSAMA 698/30/2017
- 70. Resources and References 1. Keras Documentation [https://keras.io] 2. Cheat Sheet [https://www.datacamp.com/community/blog/keras-cheat-sheet] 3. Startup.ML Deep Learning Conference: François Chollet on Keras [https://www.youtube.com/watch?v=YimQOpSRULY] 4. Machine Learning Mastery Blog [https://machinelearningmastery.com] DEEP LEARNING USING KERAS - ALY OSAMA 708/30/2017
- 71. Questions DEEP LEARNING USING KERAS - ALY OSAMA 718/30/2017
- 72. Thank You! 72















![Keras in 30 Seconds (2)
model.compile(loss='categorical_crossentropy',
optimizer='sgd',
metrics=['accuracy'])
model.compile(loss=keras.losses.categorical_crossentropy,
optimizer=keras.optimizers.SGD(lr=0.01, momentum=0.9))
2. Compile Model
Also, you can further configure your optimizer
DEEP LEARNING USING KERAS - ALY OSAMA 168/30/2017](https://image.slidesharecdn.com/kerasdeeplearning-170829192640/85/Deep-Learning-using-Keras-16-320.jpg)




![Text (20%) – Wow very Huge
0||Cyclin-dependent kinases (CDKs) regulate a variety of fundamental cellular processes. CDK10 stands out as one of the last orphan CDKs for which no activating cyclin has been identified and no kinase activity revealed. Previous work has shown that CDK10 silencing increases ETS2 (v-ets erythroblastosis virus E26
oncogene homolog 2)-driven activation of the MAPK pathway, which confers tamoxifen resistance to breast cancer cells. The precise mechanisms by which CDK10 modulates ETS2 activity, and more generally the functions of CDK10, remain elusive. Here we demonstrate that CDK10 is a cyclin-dependent kinase by
identifying cyclin M as an activating cyclin. Cyclin M, an orphan cyclin, is the product of FAM58A, whose mutations cause STAR syndrome, a human developmental anomaly whose features include toe syndactyly, telecanthus, and anogenital and renal malformations. We show that STAR syndrome-associated cyclin M
mutants are unable to interact with CDK10. Cyclin M silencing phenocopies CDK10 silencing in increasing c-Raf and in conferring tamoxifen resistance to breast cancer cells. CDK10/cyclin M phosphorylates ETS2 in vitro, and in cells it positively controls ETS2 degradation by the proteasome. ETS2 protein levels are increased
in cells derived from a STAR patient, and this increase is attributable to decreased cyclin M levels. Altogether, our results reveal an additional regulatory mechanism for ETS2, which plays key roles in cancer and development. They also shed light on the molecular mechanisms underlying STAR syndrome.Cyclin-dependent
kinases (CDKs) play a pivotal role in the control of a number of fundamental cellular processes (1). The human genome contains 21 genes encoding proteins that can be considered as members of the CDK family owing to their sequence similarity with bona fide CDKs, those known to be activated by cyclins (2). Although
discovered almost 20 y ago (3, 4), CDK10 remains one of the two CDKs without an identified cyclin partner. This knowledge gap has largely impeded the exploration of its biological functions. CDK10 can act as a positive cell cycle regulator in some cells (5, 6) or as a tumor suppressor in others (7, 8). CDK10 interacts with
the ETS2 (v-ets erythroblastosis virus E26 oncogene homolog 2) transcription factor and inhibits its transcriptional activity through an unknown mechanism (9). CDK10 knockdown derepresses ETS2, which increases the expression of the c-Raf protein kinase, activates the MAPK pathway, and induces resistance of MCF7
cells to tamoxifen (6).Here, we deorphanize CDK10 by identifying cyclin M, the product of FAM58A, as a binding partner. Mutations in this gene that predict absence or truncation of cyclin M are associated with STAR syndrome, whose features include toe syndactyly, telecanthus, and anogenital and renal malformations in
heterozygous females (10). However, both the functions of cyclin M and the pathogenesis of STAR syndrome remain unknown. We show that a recombinant CDK10/cyclin M heterodimer is an active protein kinase that phosphorylates ETS2 in vitro. Cyclin M silencing phenocopies CDK10 silencing in increasing c-Raf and
phospho-ERK expression levels and in inducing tamoxifen resistance in estrogen receptor (ER)+ breast cancer cells. We show that CDK10/cyclin M positively controls ETS2 degradation by the proteasome, through the phosphorylation of two neighboring serines. Finally, we detect an increased ETS2 expression level in cells
derived from a STAR patient, and we demonstrate that it is attributable to the decreased cyclin M expression level observed in these cells.Previous SectionNext SectionResultsA yeast two-hybrid (Y2H) screen unveiled an interaction signal between CDK10 and a mouse protein whose C-terminal half presents a strong
sequence homology with the human FAM58A gene product [whose proposed name is cyclin M (11)]. We thus performed Y2H mating assays to determine whether human CDK10 interacts with human cyclin M (Fig. 1 A–C). The longest CDK10 isoform (P1) expressed as a bait protein produced a strong interaction phenotype
with full-length cyclin M (expressed as a prey protein) but no detectable phenotype with cyclin D1, p21 (CIP1), and Cdi1 (KAP), which are known binding partners of other CDKs (Fig. 1B). CDK1 and CDK3 also produced Y2H signals with cyclin M, albeit notably weaker than that observed with CDK10 (Fig. 1B). An interaction
phenotype was also observed between full-length cyclin M and CDK10 proteins expressed as bait and prey, respectively (Fig. S1A). We then tested different isoforms of CDK10 and cyclin M originating from alternative gene splicing, and two truncated cyclin M proteins corresponding to the hypothetical products of two
mutated FAM58A genes found in STAR syndrome patients (10). None of these shorter isoforms produced interaction phenotypes (Fig. 1 A and C and Fig. S1A).Fig. 1.In a new window Download PPTFig. 1.CDK10 and cyclin M form an interaction complex. (A) Schematic representation of the different protein isoforms
analyzed by Y2H assays. Amino acid numbers are indicated. Black boxes indicate internal deletions. The red box indicates a differing amino acid sequence compared with CDK10 P1. (B) Y2H assay between a set of CDK proteins expressed as baits (in fusion to the LexA DNA binding domain) and CDK interacting proteins
expressed as preys (in fusion to the B42 transcriptional activator). pEG202 and pJG4-5 are the empty bait and prey plasmids expressing LexA and B42, respectively. lacZ was used as a reporter gene, and blue yeast are indicative of a Y2H interaction phenotype. (C) Y2H assay between the different CDK10 and cyclin M
isoforms. The amino-terminal region of ETS2, known to interact with CDK10 (9), was also assayed. (D) Western blot analysis of Myc-CDK10 (wt or kd) and CycM-V5-6His expression levels in transfected HEK293 cells. (E) Western blot analysis of Myc-CDK10 (wt or kd) immunoprecipitates obtained using the anti-Myc
antibody. “Inputs” correspond to 10 μg total lysates obtained from HEK293 cells coexpressing Myc-CDK10 (wt or kd) and CycM-V5-6His. (F) Western blot analysis of immunoprecipitates obtained using the anti-CDK10 antibody or a control goat antibody, from human breast cancer MCF7 cells. “Input” corresponds to 30 μg
MCF7 total cell lysates. The lower band of the doublet observed on the upper panel comigrates with the exogenously expressed untagged CDK10 and thus corresponds to endogenous CDK10. The upper band of the doublet corresponds to a nonspecific signal, as demonstrated by it insensitivity to either overexpression of
CDK10 (as seen on the left lane) or silencing of CDK10 (Fig. S2B). Another experiment with a longer gel migration is shown in Fig. S1D.Next we examined the ability of CDK10 and cyclin M to interact when expressed in human cells (Fig. 1 D and E). We tested wild-type CDK10 (wt) and a kinase dead (kd) mutant bearing a
D181A amino acid substitution that abolishes ATP binding (12). We expressed cyclin M-V5-6His and/or Myc-CDK10 (wt or kd) in a human embryonic kidney cell line (HEK293). The expression level of cyclin M-V5-6His was significantly increased upon coexpression with Myc-CDK10 (wt or kd) and, to a lesser extent, that of
Myc-CDK10 (wt or kd) was increased upon coexpression with cyclin M-V5-6His (Fig. 1D). We then immunoprecipitated Myc-CDK10 proteins and detected the presence of cyclin M in the CDK10 (wt) and (kd) immunoprecipitates only when these proteins were coexpressed pair-wise (Fig. 1E). We confirmed these
observations by detecting the presence of Myc-CDK10 in cyclin M-V5-6His immunoprecipitates (Fig. S1B). These experiments confirmed the lack of robust interaction between the CDK10.P2 isoform and cyclin M (Fig. S1C). To detect the interaction between endogenous proteins, we performed immunoprecipitations on
nontransfected MCF7 cells derived from a human breast cancer. CDK10 and cyclin M antibodies detected their cognate endogenous proteins by Western blotting. We readily detected cyclin M in immunoprecipitates obtained with the CDK10 antibody but not with a control antibody (Fig. 1F). These results confirm the
physical interaction between CDK10 and cyclin M in human cells.To unveil a hypothesized CDK10/cyclin M protein kinase activity, we produced GST-CDK10 and StrepII-cyclin M fusion proteins in insect cells, either individually or in combination. We observed that GST-CDK10 and StrepII-cyclin M copurified, thus confirming
their interaction in yet another cellular model (Fig. 2A). We then performed in vitro kinase assays with purified proteins, using histone H1 as a generic substrate. Histone H1 phosphorylation was detected only from lysates of cells coexpressing GST-CDK10 and StrepII-cyclin M. No phosphorylation was detected when GST-
CDK10 or StrepII-cyclin M were expressed alone, or when StrepII-cyclin M was coexpressed with GST-CDK10(kd) (Fig. 2A). Next we investigated whether ETS2, which is known to interact with CDK10 (9) (Fig. 1C), is a phosphorylation substrate of CDK10/cyclin M. We detected strong phosphorylation of ETS2 by the GST-
CDK10/StrepII-cyclin M purified heterodimer, whereas no phosphorylation was detected using GST-CDK10 alone or GST-CDK10(kd)/StrepII-cyclin M heterodimer (Fig. 2B).Fig. 2.In a new window Download PPTFig. 2.CDK10 is a cyclin M-dependent protein kinase. (A) In vitro protein kinase assay on histone H1. Lysates from
insect cells expressing different proteins were purified on a glutathione Sepharose matrix to capture GST-CDK10(wt or kd) fusion proteins alone, or in complex with STR-CycM fusion protein. Purified protein expression levels were analyzed by Western blots (Top and Upper Middle). The kinase activity was determined by
autoradiography of histone H1, whose added amounts were visualized by Coomassie staining (Lower Middle and Bottom). (B) Same as in A, using purified recombinant 6His-ETS2 as a substrate.CDK10 silencing has been shown to increase ETS2-driven c-RAF transcription and to activate the MAPK pathway (6). We
investigated whether cyclin M is also involved in this regulatory pathway. To aim at a highly specific silencing, we used siRNA pools (mix of four different siRNAs) at low final concentration (10 nM). Both CDK10 and cyclin M siRNA pools silenced the expression of their cognate targets (Fig. 3 A and C and Fig. S2) and,
interestingly, the cyclin M siRNA pool also caused a marked decrease in CDK10 protein level (Fig. 3A and Fig. S2B). These results, and those shown in Fig. 1D, suggest that cyclin M binding stabilizes CDK10. Cyclin M silencing induced an increase in c-Raf protein and mRNA levels (Fig. 3 B and C) and in phosphorylated ERK1
and ERK2 protein levels (Fig. S3B), similarly to CDK10 silencing. As expected from these effects (6), CDK10 and cyclin M silencing both decreased the sensitivity of ER+ MCF7 cells to tamoxifen, to a similar extent. The combined silencing of both genes did not result in a higher resistance to the drug (Fig. S3C). Altogether,
these observations demonstrate a functional interaction between cyclin M and CDK10, which negatively controls ETS2.Fig. 3.In a new window Download PPTFig. 3.Cyclin M silencing up-regulates c-Raf expression. (A) Western blot analysis of endogenous CDK10 and cyclin M expression levels in MCF7 cells, in response to
siRNA-mediated gene silencing. (B) Western blot analysis of endogenous c-Raf expression levels in MCF7 cells, in response to CDK10 or cyclin M silencing. A quantification is shown in Fig. S3A. (C) Quantitative RT-PCR analysis of CDK10, cyclin M, and c-Raf mRNA levels, in response to CDK10 (Upper) or cyclin M (Lower)
silencing. **P ≤ 0.01; ***P ≤ 0.001.We then wished to explore the mechanism by which CDK10/cyclin M controls ETS2. ETS2 is a short-lived protein degraded by the proteasome (13). A straightforward hypothesis is that CDK10/cyclin M positively controls ETS2 degradation. We thus examined the impact of CDK10 or cyclin
M silencing on ETS2 expression levels. The silencing of CDK10 and that of cyclin M caused an increase in the expression levels of an exogenously expressed Flag-ETS2 protein (Fig. S4A), as well as of the endogenous ETS2 protein (Fig. 4A). This increase is not attributable to increased ETS2 mRNA levels, which marginally
fluctuated in response to CDK10 or cyclin M silencing (Fig. S4B). We then examined the expression levels of the Flag-tagged ETS2 protein when expressed alone or in combination with Myc-CDK10 or -CDK10(kd), with or without cyclin M-V5-6His. Flag-ETS2 was readily detected when expressed alone or, to a lesser extent,
when coexpressed with CDK10(kd). However, its expression level was dramatically decreased when coexpressed with CDK10 alone, or with CDK10 and cyclin M (Fig. 4B). These observations suggest that endogenous cyclin M levels are in excess compared with those of CDK10 in MCF7 cells, and they show that the major
decrease in ETS2 levels observed upon CDK10 coexpression involves CDK10 kinase activity. Treatment of cells coexpressing Flag-ETS2, CDK10, and cyclin M with the proteasome inhibitor MG132 largely rescued Flag-ETS2 expression levels (Fig. 4B).Fig. 4.In a new window Download PPTFig. 4.CDK10/cyclin M controls ETS2
stability in human cancer derived cells. (A) Western blot analysis of endogenous ETS2 expression levels in MCF7 cells, in response to siRNA-mediated CDK10 and/or cyclin M silencing. A quantification is shown in Fig. S4B. (B) Western blot analysis of exogenously expressed Flag-ETS2 protein levels in MCF7 cells
cotransfected with empty vectors or coexpressing Myc-CDK10 (wt or kd), or Myc-CDK10/CycM-V5-6His. The latter cells were treated for 16 h with the MG132 proteasome inhibitor. Proper expression of CDK10 and cyclin M tagged proteins was verified by Western blot analysis. (C and D) Western blot analysis of
expression levels of exogenously expressed Flag-ETS2 wild-type or mutant proteins in MCF7 cells, in the absence of (C) or in response to (D) Myc-CDK10/CycM-V5-6His expression. Quantifications are shown in Fig. S4 C and D.A mass spectrometry analysis of recombinant ETS2 phosphorylated by CDK10/cyclin M in vitro
revealed the existence of multiple phosphorylated residues, among which are two neighboring phospho-serines (at positions 220 and 225) that may form a phosphodegron (14) (Figs. S5–S8). To confirm this finding, we compared the phosphorylation level of recombinant ETS2wt with that of ETS2SASA protein, a mutant
bearing alanine substitutions of these two serines. As expected from the existence of multiple phosphorylation sites, we detected a small but reproducible, significant decrease of phosphorylation level of ETS2SASA compared with ETS2wt (Fig. S9), thus confirming that Ser220/Ser225 are phosphorylated by CDK10/cyclin
M. To establish a direct link between ETS2 phosphorylation by CDK10/cyclin M and degradation, we examined the expression levels of Flag-ETS2SASA. In the absence of CDK10/cyclin M coexpression, it did not differ significantly from that of Flag-ETS2. This is contrary to that of Flag-ETS2DBM, bearing a deletion of the N-
terminal destruction (D-) box that was previously shown to be involved in APC-Cdh1–mediated degradation of ETS2 (13) (Fig. 4C). However, contrary to Flag-ETS2 wild type, the expression level of Flag-ETS2SASA remained insensitive to CDK10/cyclin M coexpression (Fig. 4D). Altogether, these results suggest that
CDK10/cyclin M directly controls ETS2 degradation through the phosphorylation of these two serines.Finally, we studied a lymphoblastoid cell line derived from a patient with STAR syndrome, bearing FAM58A mutation c.555+1G>A, predicted to result in aberrant splicing (10). In ……………….
8/30/2017 DEEP LEARNING USING KERAS - ALY OSAMA 21](https://image.slidesharecdn.com/kerasdeeplearning-170829192640/85/Deep-Learning-using-Keras-21-320.jpg)









![2. Keras Models - Sequential Models
1. Sequential Models : stack of layers
data_dim = 16
timesteps = 8
model = Sequential()
model.add(LSTM(32, return_sequences=True,
input_shape=(timesteps, data_dim)))
model.add(LSTM(32, return_sequences=True))
model.add(LSTM(32))
model.add(Dense(10, activation='softmax'))
model.compile(loss='categorical_crossentropy
', optimizer='rmsprop',
metrics=['accuracy'])
DEEP LEARNING USING KERAS - ALY OSAMA 318/30/2017](https://image.slidesharecdn.com/kerasdeeplearning-170829192640/85/Deep-Learning-using-Keras-31-320.jpg)











![7.2 Drop-Based Learning Rate
LearningRateScheduler: a function that takes an epoch index as input (integer, indexed from
0) and returns a new learning rate as output (float).
# learning rate schedule
def step_decay(epoch):
initial_lrate = 0.1
drop = 0.5
epochs_drop = 10.0
lrate = initial_lrate * math.pow(drop, math.floor((1+epoch)/epochs_drop))
return lrate
lrate = LearningRateScheduler(step_decay)
callbacks_list = [lrate]
# Fit the model
model.fit(X, Y, validation_split=0.33, epochs=50, batch_size=28, callbacks=callbacks_list, verbose=2)
DEEP LEARNING USING KERAS - ALY OSAMA 438/30/2017](https://image.slidesharecdn.com/kerasdeeplearning-170829192640/85/Deep-Learning-using-Keras-43-320.jpg)

![8. Metrics
1. Accuracy
• binary_accuracy
• categorical_accuracy
• sparse_categorical_accuracy
• top_k_categorical_accuracy
• sparse_top_k_categorical_accuracy
2. Precision
3. Recall
4. FScore
from keras import metrics
model.compile(loss='categorical_crossentropy',
optimizer='adadelta’,
metrics=['accuracy', 'f1score', 'precision', 'recall'])
1280/5640 [=====>........................] - ETA: 20s - loss: 1.5566 - fmeasure: 0.8134 - precision: 0.8421 - recall: 0.7867
DEEP LEARNING USING KERAS - ALY OSAMA 458/30/2017](https://image.slidesharecdn.com/kerasdeeplearning-170829192640/85/Deep-Learning-using-Keras-45-320.jpg)
![8. Metrics - Custom
Custom metrics can be passed at the compilation step. The function would need to
take (y_true, y_pred) as arguments and return a single tensor value.
import keras.backend as K
def mean_pred(y_true, y_pred):
return K.mean(y_pred)
model.compile(optimizer='rmsprop', loss='binary_crossentropy',
metrics=['accuracy', mean_pred])
DEEP LEARNING USING KERAS - ALY OSAMA 468/30/2017](https://image.slidesharecdn.com/kerasdeeplearning-170829192640/85/Deep-Learning-using-Keras-46-320.jpg)

![9.1 Automatic Verification Dataset
Keras can separate a portion of your training data into a validation dataset and evaluate the
performance of your model on that validation dataset each epoch
model.fit(X, Y, validation_split=0.33, epochs=150, batch_size=10)
...
Epoch 148/150
514/514 [==============================] - 0s - loss: 0.5219 - acc: 0.7354 - val_loss: 0.5414 - val_acc: 0.7520
Epoch 149/150
514/514 [==============================] - 0s - loss: 0.5089 - acc: 0.7432 - val_loss: 0.5417 - val_acc: 0.7520
Epoch 150/150
514/514 [==============================] - 0s - loss: 0.5148 - acc: 0.7490 - val_loss: 0.5549 - val_acc: 0.7520
DEEP LEARNING USING KERAS - ALY OSAMA 488/30/2017](https://image.slidesharecdn.com/kerasdeeplearning-170829192640/85/Deep-Learning-using-Keras-48-320.jpg)

![9.3 Manual k-Fold Cross Validation
from sklearn.model_selection import StratifiedKFold
kfold = StratifiedKFold(n_splits=10, shuffle=True, random_state=seed)
cvscores = []
for train, test in kfold.split(X, Y):
# create model
model = Sequential()
model.add(Dense(12, input_dim=8, activation='relu'))
model.add(Dense(8, activation='relu'))
model.add(Dense(1, activation='sigmoid'))
model.compile(loss='binary_crossentropy', optimizer='adam', metrics=['accuracy'])
# Fit the model
model.fit(X[train], Y[train], epochs=150, batch_size=10, verbose=0)
# evaluate the model
scores = model.evaluate(X[test], Y[test], verbose=0)
print("%s: %.2f%%" % (model.metrics_names[1], scores[1]*100))
cvscores.append(scores[1] * 100)
print("%.2f%% (+/- %.2f%%)" % (numpy.mean(cvscores), numpy.std(cvscores)))
acc: 77.92%
acc: 68.83%
acc: 72.73%
acc: 64.94%
acc: 77.92%
acc: 35.06%
acc: 74.03%
acc: 68.83%
acc: 34.21%
acc: 72.37%
64.68% (+/- 15.50%)
DEEP LEARNING USING KERAS - ALY OSAMA 508/30/2017](https://image.slidesharecdn.com/kerasdeeplearning-170829192640/85/Deep-Learning-using-Keras-50-320.jpg)


















![Resources and References
1. Keras Documentation [https://keras.io]
2. Cheat Sheet [https://www.datacamp.com/community/blog/keras-cheat-sheet]
3. Startup.ML Deep Learning Conference: François Chollet on Keras
[https://www.youtube.com/watch?v=YimQOpSRULY]
4. Machine Learning Mastery Blog [https://machinelearningmastery.com]
DEEP LEARNING USING KERAS - ALY OSAMA 708/30/2017](https://image.slidesharecdn.com/kerasdeeplearning-170829192640/85/Deep-Learning-using-Keras-70-320.jpg)

