RNA-seq: A High-resolution View of the Transcriptome
- 1. Sean Davis, M.D., Ph.D. Genetics Branch, Center for Cancer Research National Cancer Institute National Institutes of Health RNA-seq: A high-resolution View of the Transcriptome
- 4. phenotype Gene Copy Number Sequence Variation Chromatin Structure and Function Gene Expression Transcriptional Regulation DNA Methylation Patient and Population Characteristics
- 11. DNA (0.1-1.0 ug) Single molecule array Sample preparation Cluster growth 5’ 5’3’ G T C A G T C A G T C A C A G T C A T C A C C T A G C G T A G T 1 2 3 7 8 94 5 6 Image acquisition Base calling T G C T A C G A T … Sequencing Illumina SBS Technology Reversible Terminator Chemistry Foundation © Illumina, Inc.http://www.illumina.com/technology/sequencing_technology.ilmn http://seqanswers.com/forums/showthread.php?t=21
- 12. Single end vs paired end sequencing Illumina Paired-end sequencingPaired-end: useful for RRBS, essential for RNA-seq, not useful for ChIP- seq
- 13. What comes out of the machine: short reads in fastq format @D3B4KKQ1_0166:8:1101:1960:2190#CGATGT/1 CTCCTGGAAAACGCTTTGGTAGATTTGGCCAGGAGCTTTCTTTTATGTAAATTG +D3B4KKQ1_0166:8:1101:1960:2190#CGATGT/1 [^^cedeefee`cghhhfcRX`_gfghf^bZbecg^eeb[caef`ef^a_`eXa @D3B4KKQ1_0166:8:1101:2154:2137#CGATGT/1 TCCANCCATGGCAAATTCCATGGCACCGTCAAGGCTGAGAACGGGAAGCTTGTC +D3B4KKQ1_0166:8:1101:2154:2137#CGATGT/1 ab_eBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBB @D3B4KKQ1_0166:8:1101:2249:2171#CGATGT/1 TACAAGTGCAGCATCAAGGAGCGAATGCTCTACTCCAGCTGCAAGAGCCGCCTC +D3B4KKQ1_0166:8:1101:2249:2171#CGATGT/1 _[_ceeec[^eeghdffffhh^efh_egfhfgeec_fbafhhhhd`caegfheh @D3B4KKQ1_0166:8:1101:2043:2187#CGATGT/1 GAAGGAGAGAAGGGGAGGAGGGCGGGGGGCACCTACTACATCGCCCTCCACATC +D3B4KKQ1_0166:8:1101:2043:2187#CGATGT/1 ^_accceg`gga`f[fgcb`Ucgfaa_LVV^[bbbbbRWW`W^Y[_[^bbbbb @D3B4KKQ1_0166:8:1101:2188:2232#CGATGT/1 GTGGCCGATTCCTGAGCTGTGTTTGAGGAGAGGGCGGAGTGCCATCTGGGTAGC +D3B4KKQ1_0166:8:1101:2188:2232#CGATGT/1 QS to int In R: as.integer(ch arToRaw(‘e')) -33
- 14. Pair end sequencing s_8_1_sequence.txt.gz s_8_2_sequence.txt.gz @D3B4KKQ1_0166:8:1101:1960:2190#CGATGT/1 CTCCTGGAAAACGCTTTGGTAGATTTGGCCAGGAGCTTTCTTTTATGTAAATTG +D3B4KKQ1_0166:8:1101:1960:2190#CGATGT/1 [^^cedeefee`cghhhfcRX`_gfghf^bZbecg^eeb[caef`ef^a_`eXa @D3B4KKQ1_0166:8:1101:2154:2137#CGATGT/1 TCCANCCATGGCAAATTCCATGGCACCGTCAAGGCTGAGAACGGGAAGCTTGTC +D3B4KKQ1_0166:8:1101:2154:2137#CGATGT/1 ab_eBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBB @D3B4KKQ1_0166:8:1101:2249:2171#CGATGT/1 TACAAGTGCAGCATCAAGGAGCGAATGCTCTACTCCAGCTGCAAGAGCCGCCTC +D3B4KKQ1_0166:8:1101:2249:2171#CGATGT/1 _[_ceeec[^eeghdffffhh^efh_egfhfgeec_fbafhhhhd`caegfheh @D3B4KKQ1_0166:8:1101:2043:2187#CGATGT/1 GAAGGAGAGAAGGGGAGGAGGGCGGGGGGCACCTACTACATCGCCCTCCACATC +D3B4KKQ1_0166:8:1101:2043:2187#CGATGT/1 ^_accceg`gga`f[fgcb`Ucgfaa_LVV^[bbbbbRWW`W^Y[_[^bbbbb @D3B4KKQ1_0166:8:1101:2188:2232#CGATGT/1 GTGGCCGATTCCTGAGCTGTGTTTGAGGAGAGGGCGGAGTGCCATCTGGGTAGC +D3B4KKQ1_0166:8:1101:2188:2232#CGATGT/1 aa_eeeeegggggihhiiifgeghfeghbgcghifiidg^dbgggeeeee`dcd @D3B4KKQ1_0166:8:1101:1960:2190#CGATGT/2 GGCATATTTAACAGCATTGAACAGAATTCTGTGTCCTGTAAAAAAATTAGCTTA +D3B4KKQ1_0166:8:1101:1960:2190#CGATGT/2 a__aaa`ce`cgcffdf_acda^ea]befffbeged`g[a`e_caaac]cb`gb @D3B4KKQ1_0166:8:1101:2154:2137#CGATGT/2 TTGAGGCTGTTGTCATACTTCTCATGGTTCACACCCATGACGAACATGGGGGCG +D3B4KKQ1_0166:8:1101:2154:2137#CGATGT/2 a__eeeeeggegefhhhiiihhhhhiieghhhghhiiffhiififhhiihegic @D3B4KKQ1_0166:8:1101:2249:2171#CGATGT/2 CGGGGTGCACCTCGTCGTAGAGGAACTCTGCCGTCAGCTCTGCCCCATCGCCAA +D3B4KKQ1_0166:8:1101:2249:2171#CGATGT/2 ^__ee__cge`cghghhfgddgfgi]ehhfffff^ec[beegidffhhfhadba @D3B4KKQ1_0166:8:1101:2043:2187#CGATGT/2 CTTAGTCTCAGTTTTCCTCCAGCAGCCTGAGGAAACTCAAAGGCACAGTTCCCA +D3B4KKQ1_0166:8:1101:2043:2187#CGATGT/2 _abeaaacg^g^eghhhhgafghhdfghfedeghfiiicfbgdHYagfeecggf @D3B4KKQ1_0166:8:1101:2188:2232#CGATGT/2 TAGGCTCAAAGTCTAACGCCAATCCCGAACCTGGGCATCTGTACACACACACAC +D3B4KKQ1_0166:8:1101:2188:2232#CGATGT/2 abbeceeegggcghiihiihhhhiifhiiiiihiiiiiiihegh`eggfebfhg … …
- 19. Overview of BAC in the Genome
- 20. Sequencing a BAC
- 22. Repeats
- 23. Repeats
- 24. Repeats are not created equal
- 25. Approaches to RNA-seq Nature Biotech (2010) 28, 421-423
- 26. Alignment
- 30. Run Time
- 31. Alignment Yield
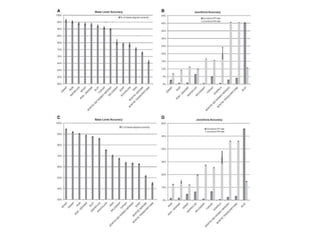
- 32. Splice Read Placement Accuracy
- 33. Impact on Transcript Assembly
- 36. Models for RNA-seq • Count-based models • Multi-reads (isoform resolution) • Paired-end reads (include length resolution step) • Positional bias along transcript length • Sequence bias
- 37. Read Counting
- 39. L. Pachter (2011) arXiv:1104.3889v
- 51. Hierarchical Clustering Gene 1 Gene 2 Gene 3 Gene 4 Gene 5 Gene 6 Gene 7 Gene 8
- 52. Hierarchical Clustering Gene 1 Gene 2 Gene 3 Gene 4 Gene 5 Gene 6 Gene 7 Gene 8
- 53. Hierarchical Clustering Gene 1 Gene 2 Gene 3 Gene 4 Gene 5 Gene 6 Gene 7 Gene 8
- 54. Hierarchical Clustering Gene 1 Gene 2 Gene 3 Gene 4 Gene 5 Gene 6 Gene 7 Gene 8
- 55. Distance Metrics Euclidean distance Manhattan distance Minkowski distance (generalized distance)
- 56. Distance Metrics • Correlation – maximum value of 1 if X and Y are perfectly correlated – minimum value of -1 if X and Y are exactly opposite – d(X,Y) = 1 – rxy • Many, many others • Choice of distance metric can be driven by underlying data (eg., binary data, categorical data, outliers, etc.)
- 57. Example of Distance Metric Choice
- 58. Example • dat = matrix(rnorm(10000),ncol=20) • dat[1:100,1:10] = dat[1:100,1:10]+1 • hclust • dist • as.dist(1-cor)
- 62. MA Plot
- 64. 64
- 65. 65
- 66. 66
- 67. 67
- 68. 68
- 71. DE False Positive Rates
- 72. DE Evaluation
- 75. RNA-seq workflow as proposed by Anders et al. in Nature Protocols
- 76. MA Plot
- 82. Fusion Detection
- 83. False Positive Fusion Detection
- 84. Experimental Design • What are my goals? – Differential expression? – Transcriptome assembly? – Identify rare, novel trancripts? • System characteristics? – Large, expanded genome? – Intron/exon structures complex? – No reference genome or transcriptome
- 85. Experimental Design • Technical replicates – Probably not needed due to low technical variation • Biological replicates – Not explicitly needed for transcript assembly – Essential for differential expression analysis – Number of replicates often driven by sample availability for human studies – More is almost always better
- 86. Links of Interest • http://bioconductor.org • http://biostars.org • http://www.rna-seqblog.com/ • https://genome.ucsc.edu/ENCODE/ • http://www.ncbi.nlm.nih.gov/gds/
Editor's Notes
- #2: I am going to spend a few minutes illustrating how existing and emerging high-throughput genomic technologies are being used to understand cancer, a mindnumbingly complex and disregulated biologic process.
- #3: The first karyotypes were produced in 1956. Shown here is a comparison of a normal karyotype of a normal female and one from a tumor. By 1960, a karyotype of a cancer genome revealed the presence of the Philadelphia chromosome. Now known to represent the BCR-ABL fusion protein, it was not until 33 years later in 1993 that a drug, gleevec, become available that targeted the fusion product. By applying high-throughput microarray technologies, the Cancer Genetics Branch is striving to make observations of the cancer genome that will provide deeper understandings of the biology of cancer, to develop prognostic and diagnostic markers to improve patient-specific treatments, and to find promising targets for directed drug therapy.
- #5: Since Knudson’s famous hypothesis proposing the two-hit model, our understanding of cancer as a genetic disease has progressed to the realization that cancer is not often a function of a single gene gone awry, but probably represents a complex interaction of multiple processes in the genome including altered copy number, gene expression, transcriptional regulation, chromatin modification, sequence variation, and DNA methylation. It is vital to the goal of producing better patient outcomes to understand not only what genes are involved in a certain type of cancer, but also how these other processes affect gene regulation. In short, an integrated view of the cancer genome is necessary and is now becoming possible.














![Pair end sequencing
s_8_1_sequence.txt.gz s_8_2_sequence.txt.gz
@D3B4KKQ1_0166:8:1101:1960:2190#CGATGT/1
CTCCTGGAAAACGCTTTGGTAGATTTGGCCAGGAGCTTTCTTTTATGTAAATTG
+D3B4KKQ1_0166:8:1101:1960:2190#CGATGT/1
[^^cedeefee`cghhhfcRX`_gfghf^bZbecg^eeb[caef`ef^a_`eXa
@D3B4KKQ1_0166:8:1101:2154:2137#CGATGT/1
TCCANCCATGGCAAATTCCATGGCACCGTCAAGGCTGAGAACGGGAAGCTTGTC
+D3B4KKQ1_0166:8:1101:2154:2137#CGATGT/1
ab_eBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBB
@D3B4KKQ1_0166:8:1101:2249:2171#CGATGT/1
TACAAGTGCAGCATCAAGGAGCGAATGCTCTACTCCAGCTGCAAGAGCCGCCTC
+D3B4KKQ1_0166:8:1101:2249:2171#CGATGT/1
_[_ceeec[^eeghdffffhh^efh_egfhfgeec_fbafhhhhd`caegfheh
@D3B4KKQ1_0166:8:1101:2043:2187#CGATGT/1
GAAGGAGAGAAGGGGAGGAGGGCGGGGGGCACCTACTACATCGCCCTCCACATC
+D3B4KKQ1_0166:8:1101:2043:2187#CGATGT/1
^_accceg`gga`f[fgcb`Ucgfaa_LVV^[bbbbbRWW`W^Y[_[^bbbbb
@D3B4KKQ1_0166:8:1101:2188:2232#CGATGT/1
GTGGCCGATTCCTGAGCTGTGTTTGAGGAGAGGGCGGAGTGCCATCTGGGTAGC
+D3B4KKQ1_0166:8:1101:2188:2232#CGATGT/1
aa_eeeeegggggihhiiifgeghfeghbgcghifiidg^dbgggeeeee`dcd
@D3B4KKQ1_0166:8:1101:1960:2190#CGATGT/2
GGCATATTTAACAGCATTGAACAGAATTCTGTGTCCTGTAAAAAAATTAGCTTA
+D3B4KKQ1_0166:8:1101:1960:2190#CGATGT/2
a__aaa`ce`cgcffdf_acda^ea]befffbeged`g[a`e_caaac]cb`gb
@D3B4KKQ1_0166:8:1101:2154:2137#CGATGT/2
TTGAGGCTGTTGTCATACTTCTCATGGTTCACACCCATGACGAACATGGGGGCG
+D3B4KKQ1_0166:8:1101:2154:2137#CGATGT/2
a__eeeeeggegefhhhiiihhhhhiieghhhghhiiffhiififhhiihegic
@D3B4KKQ1_0166:8:1101:2249:2171#CGATGT/2
CGGGGTGCACCTCGTCGTAGAGGAACTCTGCCGTCAGCTCTGCCCCATCGCCAA
+D3B4KKQ1_0166:8:1101:2249:2171#CGATGT/2
^__ee__cge`cghghhfgddgfgi]ehhfffff^ec[beegidffhhfhadba
@D3B4KKQ1_0166:8:1101:2043:2187#CGATGT/2
CTTAGTCTCAGTTTTCCTCCAGCAGCCTGAGGAAACTCAAAGGCACAGTTCCCA
+D3B4KKQ1_0166:8:1101:2043:2187#CGATGT/2
_abeaaacg^g^eghhhhgafghhdfghfedeghfiiicfbgdHYagfeecggf
@D3B4KKQ1_0166:8:1101:2188:2232#CGATGT/2
TAGGCTCAAAGTCTAACGCCAATCCCGAACCTGGGCATCTGTACACACACACAC
+D3B4KKQ1_0166:8:1101:2188:2232#CGATGT/2
abbeceeegggcghiihiihhhhiifhiiiiihiiiiiiihegh`eggfebfhg
… …](https://image.slidesharecdn.com/rnaseqbasic-160410233954/85/RNA-seq-A-High-resolution-View-of-the-Transcriptome-14-320.jpg)










































![Example
• dat = matrix(rnorm(10000),ncol=20)
• dat[1:100,1:10] = dat[1:100,1:10]+1
• hclust
• dist
• as.dist(1-cor)](https://image.slidesharecdn.com/rnaseqbasic-160410233954/85/RNA-seq-A-High-resolution-View-of-the-Transcriptome-58-320.jpg)





























