Unsupervised learning Algorithms and Assumptions
- 1. UNIT II Unsupervised Learning Dr. T. Veeramakali Assoc. Prof. / DSBS SRM Institute of Science & Technology Kattankulathur, Chennai-603203.
- 2. Topics : Introduction to unsupervised learning Unsupervised learning Algorithms and Assumptions K-Means algorithm – introduction Implementation of K-means algorithm Hierarchical Clustering – need and importance of hierarchical clustering Agglomerative Hierarchical Clustering Working of dendrogram Steps for implementation of AHC using Python Gaussian Mixture Models – Introduction, importance and need of the model Normal , Gaussian distribution Implementation of Gaussian mixture model Understand the different distance metrics used in clustering Euclidean, Manhattan, Cosine, Mahala Nobis Features of a Cluster – Labels, Centroids, Inertia, Eigen vectors and Eigen values Principal component analysis Intro to Machine Learning 2
- 3. Supervised vs. Unsupervised Learning Supervised learning (classification) Supervision: The training data (observations, measurements, etc.) are accompanied by labels indicating the class of the observations New data is classified based on the training set Unsupervised learning (clustering) The class labels of training data is unknown Given a set of measurements, observations, etc. with the aim of establishing the existence of classes or clusters in the data
- 4. Issues: Data Preparation Data cleaning Preprocess data in order to reduce noise and handle missing values Relevance analysis (feature selection) Remove the irrelevant or redundant attributes Data transformation Generalize and/or normalize data
- 5. Unsupervised Machine Learning Unsupervised Machine Learning is a technique that teaches machines to use unlabeled or unclassified data. The idea is to expose computers to large volumes of varying data and allow them to learn from that data to provide previously unknown insights and identify hidden patterns.
- 6. Unsupervised Machine Learning Unsupervised learning models utilize three main algorithms: clustering, association, and dimensionality reduction.
- 7. Unsupervised Machine Learning Clustering is the process of grouping similar entities together by categories. Unsupervised Machine Learning algorithms aim to find similarities in the data points of the dataset and group them to give us insights into the underlying patterns of different groups, for example, grouping documents by similar topics.
- 8. Start with practical applications
- 9. Start with practical applications
- 10. Start with practical applications
- 11. Start with practical applications
- 12. What is Cluster Analysis? Cluster: a collection of data objects Similar to one another within the same cluster Dissimilar to the objects in other clusters Cluster analysis Finding similarities between data according to the characteristics found in the data and grouping similar data objects into clusters Unsupervised learning: no predefined classes Typical applications As a stand-alone tool to get insight into data distribution As a preprocessing step for other algorithms
- 13. Quality: What Is Good Clustering? A good clustering method will produce high quality clusters with high intra-class similarity low inter-class similarity The quality of a clustering result depends on both the similarity measure used by the method and its implementation The quality of a clustering method is also measured by its ability to discover some or all of the hidden patterns
- 14. Measure the Quality of Clustering Dissimilarity/Similarity metric: Similarity is expressed in terms of a distance function, typically metric: d(i, j) There is a separate “quality” function that measures the “goodness” of a cluster. The definitions of distance functions are usually very different for interval-scaled, boolean, categorical, ordinal ratio, and vector variables. Weights should be associated with different variables based on applications and data semantics. It is hard to define “similar enough” or “good enough” the answer is typically highly subjective.
- 15. Typical Alternatives to Calculate the Distance between Clusters Single link: smallest distance between an element in one cluster and an element in the other, i.e., dis(Ki, Kj) = min(tip, tjq) Complete link: largest distance between an element in one cluster and an element in the other, i.e., dis(Ki, Kj) = max(tip, tjq) Average: avg distance between an element in one cluster and an element in the other, i.e., dis(Ki, Kj) = avg(tip, tjq) Centroid: distance between the centroids of two clusters, i.e., dis(Ki, Kj) = dis(Ci, Cj) Medoid: distance between the medoids of two clusters, i.e., dis(Ki, Kj) = dis(Mi, Mj) Medoid: one chosen, centrally located object in the cluster
- 16. Centroid, Radius and Diameter of a Cluster (for numerical data sets) Centroid: the “middle” of a cluster N t N i ip m C ) ( 1
- 17. Major Clustering Approaches (I) Partitioning approach: Construct various partitions and then evaluate them by some criterion, e.g., minimizing the sum of square errors Typical methods: k-means, k-medoids, CLARANS Hierarchical approach: Create a hierarchical decomposition of the set of data (or objects) using some criterion Typical methods: Diana, Agnes, BIRCH, ROCK, CAMELEON Density-based approach: Based on connectivity and density functions Typical methods: DBSACN, OPTICS, DenClue
- 18. Partitioning Algorithms: Basic Concept Partitioning method: Construct a partition of a database D of n objects into a set of k clusters, s.t., min sum of squared distance Given a k, find a partition of k clusters that optimizes the chosen partitioning criterion Global optimal: exhaustively enumerate all partitions Heuristic methods: k-means and k-medoids algorithms k-means (MacQueen’67): Each cluster is represented by the center of the cluster 2 1 ) ( mi m Km t k m t C mi
- 19. K-Means Clustering- K-Means clustering is an unsupervised iterative clustering technique. • It partitions the given data set into k predefined distinct clusters. • A cluster is defined as a collection of data points exhibiting certain similarities. It partitions the data set such that- • Each data point belongs to a cluster with the nearest mean. • Data points belonging to one cluster have high degree of similarity. • Data points belonging to different clusters have high degree of dissimilarity. The K-Means Clustering Method
- 20. The K-Means Clustering Method Given k, the k-means algorithm is implemented in four steps: Partition objects into k nonempty subsets Compute seed points as the centroids of the clusters of the current partition (the centroid is the center, i.e., mean point, of the cluster) Assign each object to the cluster with the nearest seed point Go back to Step 2, stop when no more new assignment
- 21. The K-Means Clustering Method Step 1: First, we need to provide the number of clusters, K, that need to be generated by this algorithm. Step 2: Next, choose K data points at random and assign each to a cluster. Briefly, categorize the data based on the number of data points. Step 3: The cluster centroids will now be computed. Step 4: Iterate the steps below until we find the ideal centroid, which is the assigning of data points to clusters that do not vary. 4.1 The sum of squared distances between data points and centroids would be calculated first. 4.2 At this point, we need to allocate each data point to the cluster that is closest to the others (centroid). 4.3 Finally, compute the centroids for the clusters by averaging all of the cluster’s data points.
- 22. The K-Means Clustering Method K-means implements the Expectation-Maximization strategy to solve the problem. The Expectation-step is used to assign data points to the nearest cluster, and the Maximization-step is used to compute the centroid of each cluster. When using the K-means algorithm, we must keep the following points in mind: It is suggested to normalize the data while dealing with clustering algorithms such as K-Means since such algorithms employ distance- based measurement to identify the similarity between data points. Because of the iterative nature of K-Means and the random initialization of centroids, K-Means may become stuck in a local optimum and fail to converge to the global optimum. As a result, it is advised to employ distinct centroids’ initializations.
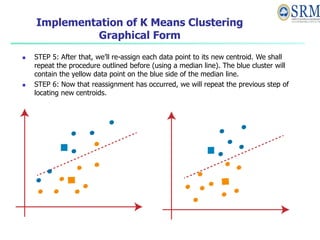
- 23. Implementation of K Means Clustering Graphical Form STEP 1: Let us pick k clusters, i.e., K=2, to separate the dataset and assign it to its appropriate clusters. We will select two random places to function as the cluster’s centroid. STEP 2: Now, each data point will be assigned to a scatter plot depending on its distance from the nearest K-point or centroid. This will be accomplished by establishing a median between both centroids. Consider the following illustration: STEP 3: The points on the line’s left side are close to the blue centroid, while the points on the line’s right side are close to the yellow centroid. The left Form cluster has a blue centroid, whereas the right Form cluster has a yellow centroid. STEP 4: Repeat the procedure, this time selecting a different centroid. To choose the new centroids, we will determine their new center of gravity, which is represented below:
- 24. Implementation of K Means Clustering Graphical Form STEP 5: After that, we’ll re-assign each data point to its new centroid. We shall repeat the procedure outlined before (using a median line). The blue cluster will contain the yellow data point on the blue side of the median line. STEP 6: Now that reassignment has occurred, we will repeat the previous step of locating new centroids.
- 25. STEP 7: We will repeat the procedure outlined above for determining the center of gravity of centroids, as shown below. STEP 8: Similar to the previous stages, we will draw the median line and reassign the data points after locating the new centroids. Implementation of K Means Clustering Graphical Form
- 26. STEP 9: We will finally group points depending on their distance from the median line, ensuring that two distinct groups are established and that no dissimilar points are included in a single group. The final Cluster is as follows: Implementation of K Means Clustering Graphical Form
- 27. Example 0 1 2 3 4 5 6 7 8 9 10 0 1 2 3 4 5 6 7 8 9 10 0 1 2 3 4 5 6 7 8 9 10 0 1 2 3 4 5 6 7 8 9 10 0 1 2 3 4 5 6 7 8 9 10 0 1 2 3 4 5 6 7 8 9 10 0 1 2 3 4 5 6 7 8 9 10 0 1 2 3 4 5 6 7 8 9 10 0 1 2 3 4 5 6 7 8 9 10 0 1 2 3 4 5 6 7 8 9 10 K=2 Arbitrarily choose K object as initial cluster center Assign each objects to most similar center Update the cluster means Update the cluster means reassign reassign Implementation of K Means Clustering Graphical Form
- 28. Distance Metrics Euclidean Distance represents the shortest distance between two points.
- 29. The K-Means Clustering Problem Use K-Means Algorithm to create two clusters-
- 30. The K-Means Clustering Problem Solution- Assume A(2, 2) and C(1, 1) are centers of the two clusters. Iteration-01: We calculate the distance of each point from each of the center of the two clusters. The distance is calculated by using the euclidean distance formula. The following illustration shows the calculation of distance between point A(2, 2) and each of the center of the two clusters-
- 31. The K-Means Clustering Problem Calculating Distance Between A(2, 2) and C1(2, 2)- Ρ(A, C1) = sqrt [ (x2 – x1)2 + (y2 – y1)2 ] = sqrt [ (2 – 2)2 + (2 – 2)2 ] = sqrt [ 0 + 0 ] = 0 Calculating Distance Between A(2, 2) and C2(1, 1)- Ρ(A, C2) = sqrt [ (x2 – x1)2 + (y2 – y1)2 ] = sqrt [ (1 – 2)2 + (1 – 2)2 ] = sqrt [ 1 + 1 ] = sqrt [ 2 ] = 1.41
- 32. The K-Means Clustering Problem From here, New clusters are- Now, We re-compute the new cluster clusters. The new cluster center is computed by taking mean of all the points contained in that cluster. This is completion of Iteration-01. Next, we go to iteration-02, iteration-03 and so on until the centers do not change anymore. Cluster-01: First cluster contains points- A(2, 2) B(3, 2) D(3, 1) Cluster-02: Second cluster contains points- C(1, 1) E(1.5, 0.5)
- 33. The K-Means Clustering Problem Iteration-2 Given Points Distance from centroid 1 (2.67,1.67) Distance from centroid 2 (1.25,0.75) Point belongs to cluster A (2,2) 0.74 1.45 C1 B(3,2) 0.33 2.15 C1 C(1,1) 1.79 0.35 C2 D(3,1) 0.74 1.76 C1 E(1.5,0.5) 1.65 0.35 C2
- 34. The K-Means Clustering Problem Iteration-2 Given Points Distance from centroid 1 (2.67,1.67) Distance from centroid 2 (1.25,0.75) Point belongs to cluster A (2,2) 0.74 1.45 C1 B(3,2) 0.33 2.15 C1 C(1,1) 1.79 0.35 C2 D(3,1) 0.74 1.76 C1 E(1.5,0.5) 1.65 0.35 C2
- 35. Clustering: Rich Applications and Multidisciplinary Efforts Pattern Recognition Spatial Data Analysis Create thematic maps in GIS by clustering feature spaces Detect spatial clusters or for other spatial mining tasks Image Processing Economic Science (especially market research) WWW Document classification Cluster Weblog data to discover groups of similar access patterns
- 36. Examples of Clustering Applications Marketing: Help marketers discover distinct groups in their customer bases, and then use this knowledge to develop targeted marketing programs Land use: Identification of areas of similar land use in an earth observation database Insurance: Identifying groups of motor insurance policy holders with a high average claim cost City-planning: Identifying groups of houses according to their house type, value, and geographical location Earth-quake studies: Observed earth quake epicenters should be clustered along continent faults
- 37. Comments on the K-Means Method Strength: Relatively efficient: O(tkn), where n is # objects, k is # clusters, and t is # iterations. Normally, k, t << n. Comment: Often terminates at a local optimum. The global optimum may be found using techniques such as: deterministic annealing and genetic algorithms Weakness Applicable only when mean is defined, then what about categorical data? Need to specify k, the number of clusters, in advance Unable to handle noisy data and outliers Not suitable to discover clusters with non-convex shapes
- 38. Hierarchical Clustering In hierarchical clustering, we assign each object (data point) to a separate cluster. Then compute the distance (similarity) between each of the clusters and join the two most similar clusters. Let’s say we have the below points and we want to cluster them into groups:
- 39. Hierarchical Clustering Let’s say we have the below points and we want to cluster them into groups: We can assign each of these points to a separate cluster:
- 40. Hierarchical Clustering Now, based on the similarity of these clusters, we can combine the most similar clusters together and repeat this process until only a single cluster is left: We are essentially building a hierarchy of clusters. That’s why this algorithm is called hierarchical clustering.
- 41. Hierarchical Clustering Types of Hierarchical Clustering There are mainly two types of hierarchical clustering: Agglomerative hierarchical clustering Divisive Hierarchical clustering
- 42. Agglomerative Hierarchical Clustering We assign each point to an individual cluster in this technique. Suppose there are 4 data points. We will assign each of these points to a cluster and hence will have 4 clusters in the beginning: Then, at each iteration, we merge the closest pair of clusters and repeat this step until only a single cluster is left:
- 43. Divisive Hierarchical Clustering Divisive hierarchical clustering works in the opposite way. Instead of starting with n clusters (in case of n observations), we start with a single cluster and assign all the points to that cluster. Now, at each iteration, we split the farthest point in the cluster and repeat this process until each cluster only contains a single point: We are splitting (or dividing) the clusters at each step, hence the name divisive hierarchical clustering.
- 44. Agglomerative Hierarchical Clustering - Example Suppose a teacher wants to divide her students into different groups. She has the marks scored by each student in an assignment and based on these marks, she wants to segment them into groups. Let’s take a sample of 5 students:
- 45. Example Creating a Proximity Matrix: First, we will create a proximity matrix which will tell us the distance between each of these points. Since we are calculating the distance of each point from each of the other points, we will get a square matrix of shape n X n (where n is the number of observations). Let’s make the 5 x 5 proximity matrix for our example: √(10-7)^2 = √9 = 3 √(10-28)^2 = √324 = 18 √(10-20)^2 = √100 = 10 √(10-35)^2 = √625 = 25 √(7-10)^2 = √9 = 3 √(7-28)^2 = √441 = 21 √(7-20)^2 = √169 = 13 √(7-35)^2 = √784 = 28
- 46. Steps to Perform Hierarchical Clustering Step 1: First, we assign all the points to an individual cluster: Step 2: Next, we will look at the smallest distance in the proximity matrix and merge the points with the smallest distance. We then update the proximity matrix: Here, the smallest distance is 3 and hence we will merge point 1 and 2.
- 47. Steps to Perform Hierarchical Clustering Let’s look at the updated clusters and accordingly update the proximity matrix: Max(7,10)=10 Now, we will again calculate the proximity matrix for these clusters:
- 48. Steps to Perform Hierarchical Clustering Step 3: We will repeat step 2 until only a single cluster is left. So, we will first look at the minimum distance in the proximity matrix and then merge the closest pair of clusters. We will get the merged clusters as shown below after repeating these steps:
- 49. Working of dendrogram A dendrogram is a diagram that shows the attribute distances between each pair of sequentially merged classes. To avoid crossing lines, the diagram is graphically arranged so that members of each pair of classes to be merged are neighbors in the diagram.
- 50. Working of dendrogram Step by step working of the algorithm: Step-1: The data points P2 and P3 merged together and form a cluster, correspondingly a dendrogram is created,. Step-2: In the next step, P5 and P6 form a cluster, and the corresponding dendrogram is created Step-3: Again, two new dendrograms are formed that combine P1, P2, and P3 in one dendrogram, and P4, P5, and P6, in another dendrogram. Step-4: At last, the final dendrogram is formed that combines all the data points together.
- 51. Hierarchical Clustering To get the number of clusters for hierarchical clustering, we make use of an awesome concept called a Dendrogram. A dendrogram is a tree-like diagram that records the sequences of merges or splits.
- 52. Hierarchical Clustering The number of clusters will be the number of vertical lines which are being intersected by the line drawn using the threshold. In the above example, since the red line intersects 2 vertical lines, we will have 2 clusters. One cluster will have a sample (1,2,4) and the other will have a sample (3,5).
- 54. Gaussian Distribution A distribution in statistics is a function that shows the possible values for a variable and how often they occur. In probability theory and statistics, the Normal Distribution, also called the Gaussian Distribution. is the most significant continuous probability distribution. Sometimes it is also called a bell curve.
- 55. Gaussian Distribution Where, x is the variable μ is the mean σ is the standard deviation Normal distribution is the most used distribution in data science. In a normal distribution graph, data is symmetrically distributed with no skew. When plotted, the data follows a bell shape, with most values clustering around a central region and tapering off as they go further away from the center.
- 65. A Gaussian mixture model is a probabilistic model that assumes all the data points are generated from a mixture of a finite number of Gaussian distributions with unknown parameters. One can think of mixture models as generalizing k-means clustering to incorporate information about the covariance structure of the data as well as the centers of the latent Gaussians.
- 66. Pros: Speed:It is the fastest algorithm for learning mixture models Agnostic:As this algorithm maximizes only the likelihood, it will not bias the means towards zero, or bias the cluster sizes to have specific structures that might or might not apply.
- 67. Cons: Singularities: When one has insufficiently many points per mixture, estimating the covariance matrices becomes difficult, and the algorithm is known to diverge and find solutions with infinite likelihood unless one regularizes the covariances artificially. Number of components:This algorithm will always use all the components it has access to, needing held-out data or information theoretical criteria to decide how many components to use in the absence of external cues.
- 68. Understand the different distance metrics used in clustering A metric or distance function is a function d(x,y) d ( x , y ) that defines the distance between elements of a set as a non-negative real number. Distance metrics are a key part of several machine learning algorithms. These distance metrics are used in both supervised and unsupervised learning, generally to calculate the similarity between data points. An effective distance metric improves the performance of our machine learning model.
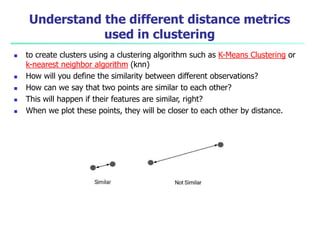
- 69. Understand the different distance metrics used in clustering to create clusters using a clustering algorithm such as K-Means Clustering or k-nearest neighbor algorithm (knn) How will you define the similarity between different observations? How can we say that two points are similar to each other? This will happen if their features are similar, right? When we plot these points, they will be closer to each other by distance.
- 70. Understand the different distance metrics used in clustering how do we calculate this distance, and what are the different distance metrics in machine learning? Also, are these metrics different for different learning problems? Do we use any special theorem for this? Types of Distance Metrics in Machine Learning Euclidean Distance Manhattan Distance Minkowski Distance Hamming Distance
- 71. Understand the different distance metrics used in clustering Euclidean Distance Euclidean Distance represents the shortest distance between two vectors. It is the square root of the sum of squares of differences between corresponding elements.
- 72. Understand the different distance metrics used in clustering Euclidean Distance Formula for Euclidean Distance
- 73. Understand the different distance metrics used in clustering Euclidean Distance We can generalize this for an n-dimensional space as: Where, •n = number of dimensions •pi, qi = data points
- 74. Understand the different distance metrics used in clustering # importing the library from scipy.spatial import distance # defining the points point_1 = (1, 2, 3) point_2 = (4, 5, 6) point_1, point_2 OUTPUT: ((1, 2, 3), (4, 5, 6))
- 75. Understand the different distance metrics used in clustering # importing the library from scipy.spatial import distance # defining the points point_1 = (1, 2, 3) point_2 = (4, 5, 6) point_1, point_2 # computing the euclidean distance euclidean_distance = distance.euclidean(point_1, point_2) print('Euclidean Distance b/w', point_1, 'and', point_2, 'is: ', euclidean_distance) OUTPUT: Euclidean Distance b/w (1, 2, 3) and (4, 5, 6) is: 5.196152422706632
- 76. Understand the different distance metrics used in clustering Manhattan Distance Manhattan Distance is the sum of absolute differences between points across all the dimensions. Formula for Manhattan Distance
- 77. Understand the different distance metrics used in clustering Manhattan Distance Manhattan Distance is the sum of absolute differences between points across all the dimensions. formula for an n-dimensional space is given as: Where, •n = number of dimensions •pi, qi = data points
- 78. Understand the different distance metrics used in clustering Manhattan Distance manhattan_distance = distance.cityblock(point_1, point_2) print('Manhattan Distance b/w', point_1, 'and', point_2, 'is: ', manhattan_distance) OUTPUT: Manhattan Distance b/w (1, 2, 3) and (4, 5, 6) is: 9
- 79. Understand the different distance metrics used in clustering Cosine Cosine similarity is the cosine of the angle between the vectors; that is, it is the dot product of the vectors divided by the product of their lengths. The cosine similarity is calculated as: The cosine distance formula is then: 1 - Cosine Similarity
- 80. Cosine Distance & Cosine Similarity: Cosine distance & Cosine Similarity metric is mainly used to find similarities between two data points. As the cosine distance between the data points increases, the cosine similarity, or the amount of similarity decreases, and vice versa. Thus, Points closer to each other are more similar than points that are far away from each other. Cosine similarity is given by Cos θ, and cosine distance is 1- Cos θ. Example:-
- 81. In the above image, there are two data points shown in blue, the angle between these points is 90 degrees, and Cos 90 = 0. Therefore, the shown two points are not similar, and their cosine distance is 1 — Cos 90 = 1.
- 82. Now if the angle between the two points is 0 degrees in the above figure, then the cosine similarity, Cos 0 = 1 and Cosine distance is 1- Cos 0 = 0. Then we can interpret that the two points are 100% similar to each other.
- 83. In the above figure, imagine the value of θ to be 60 degrees, then by cosine similarity formula, Cos 60 =0.5 and Cosine distance is 1- 0.5 = 0.5. Therefore the points are 50% similar to each other.
- 84. Understand the different distance metrics used in clustering Cosine Distance: Let's use an example to calculate the similarity between two fruits – strawberries (vector A) and blueberries (vector B). Since our data is already represented as a vector, we can calculate the distance. Strawberry → [4,0,1] Blueberry → [3,0,1]
- 85. The distance can be calculated using the below formula:- Minkowski distance is a generalized distance metric. We can manipulate the above formula by substituting ‘p’ to calculate the distance between two data points in different ways. Thus, Minkowski Distance is also known as Lp norm distance. Some common values of ‘p’ are:- •p = 1, Manhattan Distance •p = 2, Euclidean Distance •p = infinity, Chebychev Distance
- 87. Understand the different distance metrics used in clustering Cosine Distance:
- 88. Mahalanobis Distance is used for calculating the distance between two data points in a multivariate space. Mahala Nobis distance takes covariance in account which helps in measuring the strength/similarity between two different data objects. 99 Mahalanobis Distance Here, S is the covariance metrics. We are using inverse of the covariance metric to get a variance-normalized distance equation.
- 89. In a regular Euclidean space, variables (e.g. x, y, z) are represented by axes drawn at right angles to each other; The distance between any two points can be measured with a ruler. For uncorrelated variables, the Euclidean distance equals the MD. However, if two or more variables are correlated, the axes are no longer at right angles, and the measurements become impossible with a ruler.
- 90. In addition, if you have more than three variables, you can’t plot them in regular 3D space at all. The MD solves this measurement problem, as it measures distances between points, even correlated points for multiple variables.
- 91. Mahalanobis distance plot example. A contour plot overlaying the scatterplot of 100 random draws from a bivariate normal distribution with mean zero, unit variance, and 50% correlation. The centroid defined by the marginal means is noted by a blue square.
- 92. The Mahalanobis distance measures distance relative to the centroid — a base or central point which can be thought of as an overall mean for multivariate data. The centroid is a point in multivariate space where all means from all variables intersect. The larger the MD, the further away from the centroid the data point is.
- 93. if the dimensions (columns in your dataset) are correlated to one another, which is typically the case in real-world datasets, the Euclidean distance between a point and the center of the points (distribution) can give little or misleading information about how close a point really is to the cluster.
- 95. Uses The most common use for the Mahalanobis distance is to find multivariate outliers, which indicates unusual combinations of two or more variables. For example, it’s fairly common to find a 6′ tall woman weighing 185 lbs, but it’s rare to find a 4′ tall woman who weighs that much.
- 96. Inertia measures how well a dataset was clustered by K- Means. It is calculated by measuring the distance between each data point and its centroid, squaring this distance, and summing these squares across one cluster. A good model is one with low inertia AND a low number of clusters (K). However, this is a tradeoff because as K increases, inertia decreases.. 107 Inertia
- 97. To find the optimal K for a dataset, use the Elbow method; find the point where the decrease in inertia begins to slow. K=3 is the “elbow” of this graph. 108 Inertia
- 98. It tells us how far the points within a cluster are. So, inertia actually calculates the sum of distances of all the points within a cluster from the centroid of that cluster. Normally, we use Euclidean distance as the distance metric, as long as most of the features are numeric; otherwise, Manhattan distance in case most of the features are categorical. We calculate this for all the clusters; the final inertial value is the sum of all these distances. This distance within the clusters is known as intracluster distance. So, inertia gives us the sum of intracluster distances:
- 99. the distance between them should be as low as possible.
- 100. Eigen vectors and Eigen values Eigenvalues and eigenvectors are concepts from linear algebra that are used to analyze and understand linear transformations, particularly those represented by square matrices. They are used in many different areas of mathematics, including machine learning and artificial intelligence. used to determine a set of important variables (in form of a vector) along with scale along different dimensions (key dimensions based on variance) for analyzing the data in a better manner.
- 101. In machine learning, eigenvalues and eigenvectors are used to represent data, to perform operations on data, and to train machine learning models. In artificial intelligence, eigenvalues and eigenvectors are used to develop algorithms for tasks such as image recognition, natural language processing, and robotics. 112 Eigen vectors and Eigen values
- 102. Eigenvalues are the special set of scalar values that is associated with the set of linear equations most probably in the matrix equations. Eigenvectors are also termed as characteristic roots. It is a non-zero vector that can be changed at most by its scalar factor after the application of linear transformations. Av=λv 113 Eigen vectors and Eigen values
- 103. 1. Eigenvalue (λ): An eigenvalue of a square matrix A is a scalar (a single number) λ such that there exists a non-zero vector v (the eigenvector) for which the following equation holds: Av = λv In other words, when you multiply the matrix A by the eigenvector v, you get a new vector that is just a scaled version of v (scaled by the eigenvalue λ).
- 104. 2. Eigenvector: The vector v mentioned above is called an eigenvector corresponding to the eigenvalue λ. Eigenvectors only change in scale (magnitude) when multiplied by the matrix A; their direction remains the same. Mathematically, to find eigenvalues and eigenvectors, you typically solve the following equation for λ and v: (A — λI)v = 0
- 105. (A — λI)v = 0 Where: A is the square matrix for which you want to find eigenvalues and eigenvectors. λ is the eigenvalue you’re trying to find. I is the identity matrix (a diagonal matrix with 1s on the diagonal and 0s elsewhere). v is the eigenvector you’re trying to find.
- 106. Eigenvectors are the vectors that when multiplied by a matrix (linear combination or transformation) result in another vector having the same direction but scaled (hence scaler multiple) in forward or reverse direction by a magnitude of the scaler multiple which can be termed as
- 107. Eigenvalue. In simpler words, the eigenvalues are scalar values that represent the scaling factor by which a vector is transformed when a linear transformation is applied. In other words, eigenvalues are the values that scale eigenvectors when a linear transformation is applied.
- 108. When the matrix multiplication with vector results in another vector in the same/opposite direction but scaled in forward / reverse direction by a magnitude of scaler multiple or eigenvalue , then the vector is called the eigenvector of that matrix. Here is the diagram representing the eigenvector x of matrix A because the vector Ax is in the same/opposite direction of x.
- 109. An eigenvector is a vector whose direction remains unchanged when a linear transformation is applied to it. Consider the image below in which three vectors are shown. The green square is only drawn to illustrate the linear transformation that is applied to each of these three vectors. Eigenvectors (red) do not change direction when a linear transformation (e.g. scaling) is applied to them. Other vectors (yellow) do.
- 111. First Eigen value
- 112. Second Eigen value We then arbitrarily choose , and find . Therefore, the eigenvector that corresponds to eigenvalue is
- 114. Use of Eigenvalues and Eigenvectors in Machine Learning and AI: 1. Dimensionality Reduction (PCA): In Principal Component Analysis (PCA), you calculate the eigenvectors and e¯igenvalues of the covariance matrix of your datˀa. The eigenvectors (principal components) with the largest eigenvalues capture the most variance in the data and can be used to reduce the dimensionality of the dataset while preserving important information. 2. Image Compression: Eigenvectors and eigenvalues are used in techniques like Singular Value Decomposition (SVD) for image compression. By representing images in terms of their eigenvectors and eigenvalues, you can reduce storage requirements while retaining essential image features.
- 115. 3. Support vector machines: Support vector machines (SVMs) are a type of machine learning algorithm that can be used for classification and regression tasks. SVMs work by finding a hyperplane that separates the data into two classes. The eigenvalues and eigenvectors of the kernel matrix of the SVM can be used to improve the performance of the algorithm. 4. Graph Theory: Eigenvectors play a role in analyzing networks and graphs. They can be used to find important nodes or communities in social networks or other interconnected systems.
- 116. 5. Natural Language Processing (NLP): In NLP, eigenvectors can help identify the most relevant terms in a large document-term matrix, enabling techniques like Latent Semantic Analysis (LSA) for document retrieval and text summarization. 6. Machine Learning Algorithms: Eigenvalues and eigenvectors can be used to analyze the stability and convergence properties of machine learning algorithms, especially in deep learning when dealing with weight matrices in neural networks.
- 117. Example 1: Principal Component Analysis (PCA) PCA is a widely used dimensionality reduction technique in machine learning and data analysis. It employs eigenvectors and eigenvalues to reduce the number of features while retaining as much information as possible. Imagine you have a dataset with two variables, X and Y, and you want to reduce it to one dimension. You calculate the covariance matrix of your data and find its eigenvectors and eigenvalues. Let’s say you obtain the following: • Eigenvalue 1 (λ₁) = 5 • Eigenvalue 2 (λ₂) = 1 • Eigenvector 1 (v₁) = [0.8, 0.6] • Eigenvector 2 (v₂) = [-0.6, 0.8] In PCA, you would choose the eigenvector corresponding to the largest eigenvalue as the principal component. In this case, it’s v₁. You project your data onto this eigenvector to reduce it to one dimension, effectively capturing most of the variance in the data.
- 118. There are several key properties of eigenvalues and eigenvectors that are important to understand. These include: 1. Eigenvectors are non-zero vectors: Eigenvectors cannot be zero vectors, as this would imply that the transformation has no effect on the vector. 2. Eigenvalues can be real or complex: Eigenvalues can be either real or complex numbers, depending on the matrix being analyzed. 3. Eigenvectors can be scaled: Eigenvectors can be scaled by any non-zero scalar value and still be valid eigenvectors. 4. Eigenvectors can be orthogonal: Eigenvectors that correspond to different eigenvalues are always orthogonal to each other.
- 119. Principal component analysis (PCA) is a dimensionality reduction and machine learning method used to simplify a large data set into a smaller set. which is the process of reducing the number of predictor variables in a dataset. PCA is an unsupervised type of feature extraction, where original variables are combined and reduced to their most important and descriptive components. The goal of PCA is to identify patterns in a data set, and then distill the variables down to their most important features so that the data is simplified without losing important traits. 130 Principal component analysis
- 120. PCA looks at the overall structure of the continuous variables in a data set to extract meaningful signals from the noise in the data set. It aims to eliminate redundancy in variables while preserving important information. 131 Principal component analysis
- 121. PCA originally hails from the field of linear algebra. It is a transformation method that creates (weighted linear) combinations of the original variables in a data set, with the intent that the new combinations will capture as much variance (i.e., the separation between points) in the dataset as possible while eliminating correlations (i.e., redundancy). PCA creates the new variables by transforming the original (mean-centered) observations (records) in a dataset to a new set of variables (dimensions) using the eigenvectors and eigenvalues calculated from a covariance matrix of your original variables. 132 Principal component analysis
- 122. Basic Terminologies of PCA Before getting into PCA, we need to understand some basic terminologies, • Variance – for calculating the variation of data distributed across dimensionality of graph • Covariance – calculating dependencies and relationship between features • Standardizing data – Scaling our dataset within a specific range for unbiased output
- 123. in scaling, you're changing the range of your data Normalization: in normalization you're changing the shape of the distribution of your data. Data is transformed into a range between 0 and 1 by normalization, which involves dividing a vector by its length. Standardization is divided by the standard deviation after the mean has been subtracted.
- 124. Covariance matrix – Used for calculating interdependencies between the features or variables and also helps in reduce it to improve the performance •Covariance matrix – Used for calculating interdependencies between the features or variables and also helps in reduce it to improve the performance
- 125. How does PCA work? The steps involved for PCA are as follows- 1. Original Data 2. Normalize the original data (mean =0, variance =1) 3. Calculating covariance matrix 4. Calculating Eigen values, Eigen vectors, and normalized Eigenvectors 5. Calculating Principal Component (PC) 6. Plot the graph for orthogonality between PCs
- 126. A variance-covariance matrix is a square matrix (has the same number of rows and columns) that gives the covariance between each pair of elements available in the data. Covariance measures the extent to which to variables move in the same direction.
- 127. A covariance matrix is: • Symmetric • The square matrix is equal to its transpose: 𝐴=𝐴𝑇. • Positive semi-definite • With main diagonal containing the variances (covariances of variables on themselves)
- 128. variance-covariance-matrix-using- python/ Variance-Covariance Matrix Example First, let’s consider some sample data to work with: Age Experience Salary 0 25 2 2000 1 32 6 3000 2 37 9 3500
- 136. Step 1: Standardization First, we need to standardize our dataset to ensure that each variable has a mean of 0 and a standard deviation of 1. Here, μ is the mean of independent features σ is the standard deviation of independent features 147 Principal component analysis
- 137. Step2: Covariance Matrix Computation Covariance measures the strength of joint variability between two or more variables, indicating how much they change in relation to each other. To find the covariance we can use the formula: The value of covariance can be positive, negative, or zeros. Positive: As the x1 increases x2 also increases. Negative: As the x1 increases x2 also decreases. Zeros: No direct relation 148 Principal component analysis
- 139. How Principal Component Analysis(PCA) works? import pandas as pd import numpy as np # Here we are using inbuilt dataset of scikit learn from sklearn.datasets import load_breast_cancer # instantiating cancer = load_breast_cancer(as_frame=True) # creating dataframe df = cancer.frame # checking shape print('Original Dataframe shape :',df.shape) # Input features X = df[cancer['feature_names']] print('Inputs Dataframe shape :', X.shape) 150 Principal component analysis
- 140. first calculate the mean and standard deviation of each feature in the feature space. # Mean X_mean = X.mean() # Standard deviation X_std = X.std() # Standardization Z = (X - X_mean) / X_std 151 Principal component analysis
- 141. The covariance matrix helps us visualize how strong the dependency of two features is with each other in the feature space. # covariance c = Z.cov() # Plot the covariance matrix import matplotlib.pyplot as plt import seaborn as sns sns.heatmap(c) plt.show() 152 Principal component analysis
- 142. Now we will compute the eigenvectors and eigenvalues for our feature space which serve a great purpose in identifying the principal components for our feature space. eigenvalues, eigenvectors = np.linalg.eig(c) print('Eigen values:n', eigenvalues) print('Eigen values Shape:', eigenvalues.shape) print('Eigen Vector Shape:', eigenvectors.shape) 153 Principal component analysis
- 143. Sort the eigenvalues in descending order and sort the corresponding eigenvectors accordingly. # Index the eigenvalues in descending order idx = eigenvalues.argsort()[::-1] # Sort the eigenvalues in descending order eigenvalues = eigenvalues[idx] # sort the corresponding eigenvectors accordingly eigenvectors = eigenvectors[:,idx] 154 Principal component analysis
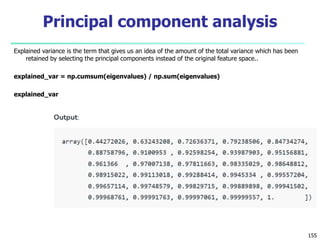
- 144. Explained variance is the term that gives us an idea of the amount of the total variance which has been retained by selecting the principal components instead of the original feature space.. explained_var = np.cumsum(eigenvalues) / np.sum(eigenvalues) explained_var 155 Principal component analysis
- 145. Determine the Number of Principal Components Here we can either consider the number of principal components of any value of our choice or by limiting the explained variance. Here considering explained variance more than equal to 50%. Let’s check how many principal components come into this. n_components = np.argmax(explained_var >= 0.50) + 1 n_components Output: 2 156 Principal component analysis
- 146. # PCA component or unit matrix u = eigenvectors[:,:n_components] pca_component = pd.DataFrame(u, index = cancer['feature_names'], columns = ['PC1','PC2'] ) # plotting heatmap plt.figure(figsize =(5, 7)) sns.heatmap(pca_component) plt.title('PCA Component') plt.show() 157 Principal component analysis
- 147. # Matrix multiplication or dot Product Z_pca = Z @ pca_component # Rename the columns name Z_pca.rename({'PC1': 'PCA1', 'PC2': 'PCA2'}, axis=1, inplace=True) # Print the Pricipal Component values print(Z_pca) 158 Principal component analysis
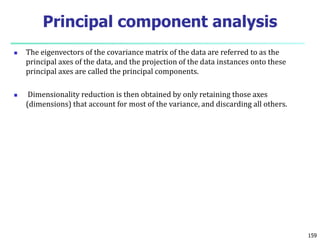
- 148. The eigenvectors of the covariance matrix of the data are referred to as the principal axes of the data, and the projection of the data instances onto these principal axes are called the principal components. Dimensionality reduction is then obtained by only retaining those axes (dimensions) that account for most of the variance, and discarding all others. 159 Principal component analysis
- 149. PCA using Using Sklearn There are different libraries in which the whole process of the principal component analysis has been automated by implementing it in a package as a function and we just have to pass the number of principal components which we would like to have. Sklearn is one such library that can be used for the PCA as shown below. # Importing PCA from sklearn.decomposition import PCA # Let's say, components = 2 pca = PCA(n_components=2) pca.fit(Z) x_pca = pca.transform(Z) # Create the dataframe df_pca1 = pd.DataFrame(x_pca, columns=['PC{}'. format(i+1) for i in range(n_components)]) print(df_pca1) 160 Principal component analysis
- 150. # giving a larger plot plt.figure(figsize=(8, 6)) plt.scatter(x_pca[:, 0], x_pca[:, 1], c=cancer['target'], cmap='plasma') # labeling x and y axes plt.xlabel('First Principal Component') plt.ylabel('Second Principal Component') plt.show() 161 Principal component analysis
- 151. # components pca.components_ 162 Principal component analysis
- 152. 163





























![The K-Means Clustering Problem
Calculating Distance Between A(2, 2) and
C1(2, 2)-
Ρ(A, C1)
= sqrt [ (x2 – x1)2 + (y2 – y1)2 ]
= sqrt [ (2 – 2)2 + (2 – 2)2 ]
= sqrt [ 0 + 0 ]
= 0
Calculating Distance Between A(2, 2)
and C2(1, 1)-
Ρ(A, C2)
= sqrt [ (x2 – x1)2 + (y2 – y1)2 ]
= sqrt [ (1 – 2)2 + (1 – 2)2 ]
= sqrt [ 1 + 1 ]
= sqrt [ 2 ]
= 1.41](https://image.slidesharecdn.com/imlunit-2-240413165241-b5a4543c/85/Unsupervised-learning-Algorithms-and-Assumptions-31-320.jpg)



















































![Understand the different distance metrics
used in clustering
Cosine Distance:
Let's use an example to calculate the similarity between two fruits –
strawberries (vector A) and blueberries (vector B). Since our data is
already represented as a vector, we can calculate the distance.
Strawberry → [4,0,1]
Blueberry → [3,0,1]](https://image.slidesharecdn.com/imlunit-2-240413165241-b5a4543c/85/Unsupervised-learning-Algorithms-and-Assumptions-84-320.jpg)
































![Example 1: Principal Component
Analysis (PCA)
PCA is a widely used dimensionality reduction technique in machine
learning and data analysis.
It employs eigenvectors and eigenvalues to reduce the number of
features while retaining as much information as possible.
Imagine you have a dataset with two variables, X and Y, and you want
to reduce it to one dimension. You calculate the covariance matrix of
your data and find its eigenvectors and eigenvalues. Let’s say you
obtain the following:
• Eigenvalue 1 (λ₁) = 5
• Eigenvalue 2 (λ₂) = 1
• Eigenvector 1 (v₁) = [0.8, 0.6]
• Eigenvector 2 (v₂) = [-0.6, 0.8]
In PCA, you would choose the eigenvector corresponding to the
largest eigenvalue as the principal component. In this case, it’s v₁. You
project your data onto this eigenvector to reduce it to one dimension,
effectively capturing most of the variance in the data.](https://image.slidesharecdn.com/imlunit-2-240413165241-b5a4543c/85/Unsupervised-learning-Algorithms-and-Assumptions-117-320.jpg)





















![ How Principal Component Analysis(PCA) works?
import pandas as pd
import numpy as np
# Here we are using inbuilt dataset of scikit learn
from sklearn.datasets import load_breast_cancer
# instantiating
cancer = load_breast_cancer(as_frame=True)
# creating dataframe
df = cancer.frame
# checking shape
print('Original Dataframe shape :',df.shape)
# Input features
X = df[cancer['feature_names']]
print('Inputs Dataframe shape :', X.shape)
150
Principal component analysis](https://image.slidesharecdn.com/imlunit-2-240413165241-b5a4543c/85/Unsupervised-learning-Algorithms-and-Assumptions-139-320.jpg)



![Sort the eigenvalues in descending order and sort the corresponding eigenvectors accordingly.
# Index the eigenvalues in descending order
idx = eigenvalues.argsort()[::-1]
# Sort the eigenvalues in descending order
eigenvalues = eigenvalues[idx]
# sort the corresponding eigenvectors accordingly
eigenvectors = eigenvectors[:,idx]
154
Principal component analysis](https://image.slidesharecdn.com/imlunit-2-240413165241-b5a4543c/85/Unsupervised-learning-Algorithms-and-Assumptions-143-320.jpg)

![# PCA component or unit matrix
u = eigenvectors[:,:n_components]
pca_component = pd.DataFrame(u,
index = cancer['feature_names'],
columns = ['PC1','PC2']
)
# plotting heatmap
plt.figure(figsize =(5, 7))
sns.heatmap(pca_component)
plt.title('PCA Component')
plt.show()
157
Principal component analysis](https://image.slidesharecdn.com/imlunit-2-240413165241-b5a4543c/85/Unsupervised-learning-Algorithms-and-Assumptions-146-320.jpg)

![ PCA using Using Sklearn
There are different libraries in which the whole process of the principal
component analysis has been automated by implementing it in a package
as a function and we just have to pass the number of principal components
which we would like to have. Sklearn is one such library that can be used
for the PCA as shown below.
# Importing PCA
from sklearn.decomposition import PCA
# Let's say, components = 2
pca = PCA(n_components=2)
pca.fit(Z)
x_pca = pca.transform(Z)
# Create the dataframe
df_pca1 = pd.DataFrame(x_pca,
columns=['PC{}'.
format(i+1)
for i in range(n_components)])
print(df_pca1)
160
Principal component analysis](https://image.slidesharecdn.com/imlunit-2-240413165241-b5a4543c/85/Unsupervised-learning-Algorithms-and-Assumptions-149-320.jpg)
![ # giving a larger plot
plt.figure(figsize=(8, 6))
plt.scatter(x_pca[:, 0], x_pca[:, 1],
c=cancer['target'],
cmap='plasma')
# labeling x and y axes
plt.xlabel('First Principal Component')
plt.ylabel('Second Principal Component')
plt.show()
161
Principal component analysis](https://image.slidesharecdn.com/imlunit-2-240413165241-b5a4543c/85/Unsupervised-learning-Algorithms-and-Assumptions-150-320.jpg)

